Module: Plotting#
One of the main usecases of SegTraQ is to compare different segmentation methods. To facilitate this task, you can make use of the plotting module (pl). In order to use it, you need to prepare your data in the form of a dictionary that maps the method name to the corresponding SegTraQ object.
To follow along with this tutorial, you can download the data from here.
Data Preparation#
First, we need to get the data into the right format. This closely follows the Xenium Focus.
[1]:
%load_ext autoreload
%autoreload 2
import warnings
import anndata as ad
import scanpy as sc
import spatialdata as sd
import segtraq
warnings.filterwarnings(action="ignore")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
[2]:
# === READING SPATIALDATA OBJECTS ===
sdata_xenium_ws = sd.read_zarr("../../data/xenium_5K_data/xenium.zarr")
sdata_bidcell_ws = sd.read_zarr("../../data/xenium_5K_data/bidcell.zarr")
sdata_proseg_ws = sd.read_zarr("../../data/xenium_5K_data/proseg2.zarr")
sdata_segger_ws = sd.read_zarr("../../data/xenium_5K_data/segger.zarr")
# === CROPPING ===
bb_xmin = 800
bb_ymin = 1150
bb_w = 200
bb_h = 300
bb_xmax = bb_xmin + bb_w
bb_ymax = bb_ymin + bb_h
def crop(ws):
return ws.query.bounding_box(
axes=["x", "y"],
min_coordinate=[bb_xmin, bb_ymin],
max_coordinate=[bb_xmax, bb_ymax],
target_coordinate_system="global",
)
sdata_xenium = crop(sdata_xenium_ws)
sdata_bidcell = crop(sdata_bidcell_ws)
sdata_proseg = crop(sdata_proseg_ws)
sdata_segger = crop(sdata_segger_ws)
# === CREATING SEGTRAQ OBJECTS ===
st_xenium = segtraq.SegTraQ(
sdata_xenium, images_key="image", tables_centroid_x_key="x_centroid", tables_centroid_y_key="y_centroid"
)
st_bidcell = segtraq.SegTraQ(
sdata_bidcell,
images_key="image",
points_background_id=0,
tables_centroid_x_key="centroid_x",
tables_centroid_y_key="centroid_y",
)
st_proseg = segtraq.SegTraQ(
sdata_proseg,
images_key="image",
points_background_id=0,
tables_area_key=None,
tables_centroid_x_key="centroid_x",
tables_centroid_y_key="centroid_y",
)
st_segger = segtraq.SegTraQ(
sdata_segger, images_key="image", tables_centroid_x_key="cell_centroid_x", tables_centroid_y_key="cell_centroid_y"
)
# === PUTTING EVERYTHING INTO A DICTIONARY ===
st_dict = {"xenium": st_xenium, "bidcell": st_bidcell, "proseg": st_proseg, "segger": st_segger}
# === REMOVING CONTROL PROBES ===
for method, st in st_dict.items():
qv = 30 if method == "segger" else 20
st.filter_control_and_low_quality_transcripts(min_qv=qv)
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd55160>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd6a0d0>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6fe7df90>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd5c8a0>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6ffe3e10>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd6f530>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6fed9e10>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6feda580>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd66c50>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6dd66e50>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6ff299a0>: None
no parent found for <ome_zarr.reader.Label object at 0x7fce6ff2b890>: None
Run desired metrics#
Now, we can run all of the metrics we want on this dataset. For now, we simply run a label transfer and run some scanpy functions to compute the UMAP coordinates.
[3]:
# running computations for UMAP etc.
for _, st in st_dict.items():
# extracting the anndata object from the spatialdata object and performing appropriate normalization
adata = st.sdata.tables["table"]
adata.layers["raw"] = adata.X.copy()
sc.pp.normalize_total(adata, inplace=True)
sc.pp.log1p(adata)
sc.pp.pca(adata)
sc.pp.neighbors(adata)
sc.tl.umap(adata)
# Load scRNA-seq data
adata_ref = ad.read_h5ad("../../data/xenium_5K_data/BC_scRNAseq_Janesick.h5ad")
# Transfer labels
for _method, st in st_dict.items():
st.run_label_transfer(
adata_ref, ref_cell_type="celltype_major", inplace=True, ref_ensemble_key=None, query_ensemble_key=None
)
Plotting#
Now, we can use the pl module to create some plots. All we need to do is pass our dictionary of SegTraQ objects.
[4]:
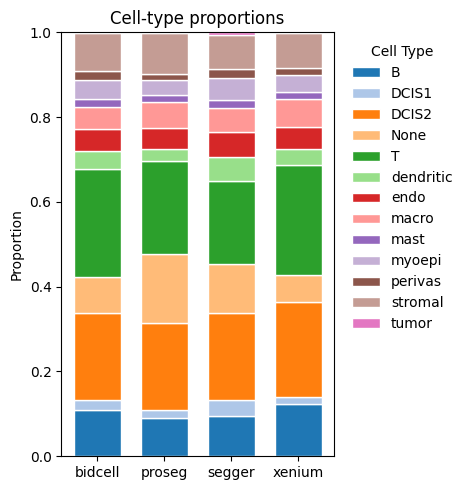
_ = segtraq.pl.celltype_proportions(st_dict, celltype_col="transferred_cell_type")
[5]:
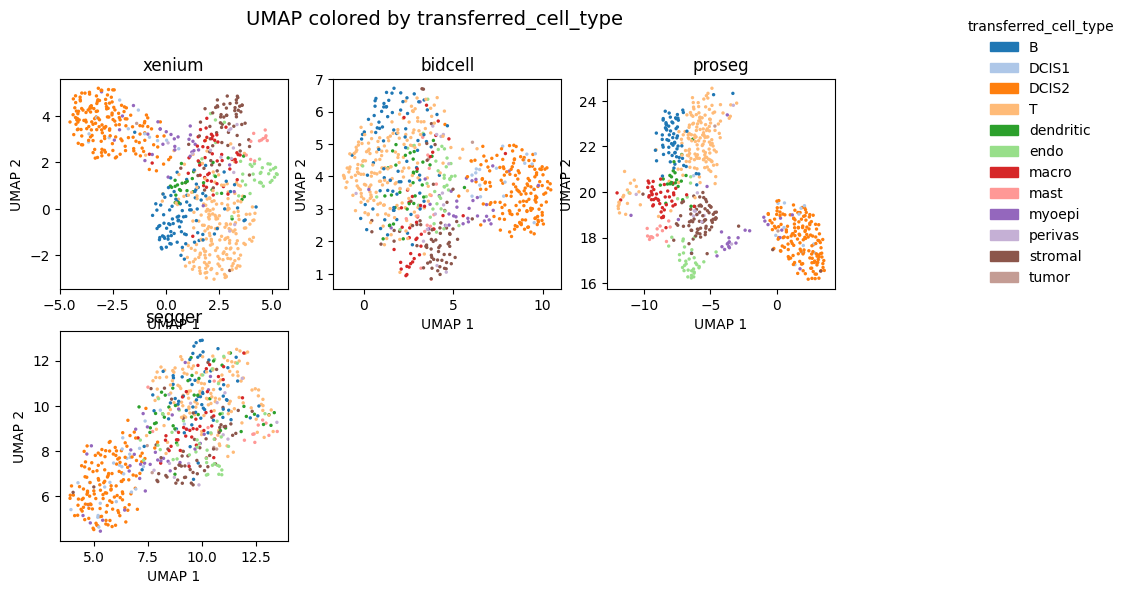
_ = segtraq.pl.umap(st_dict, color="transferred_cell_type", legend=True, figsize=(10, 6))
[6]:
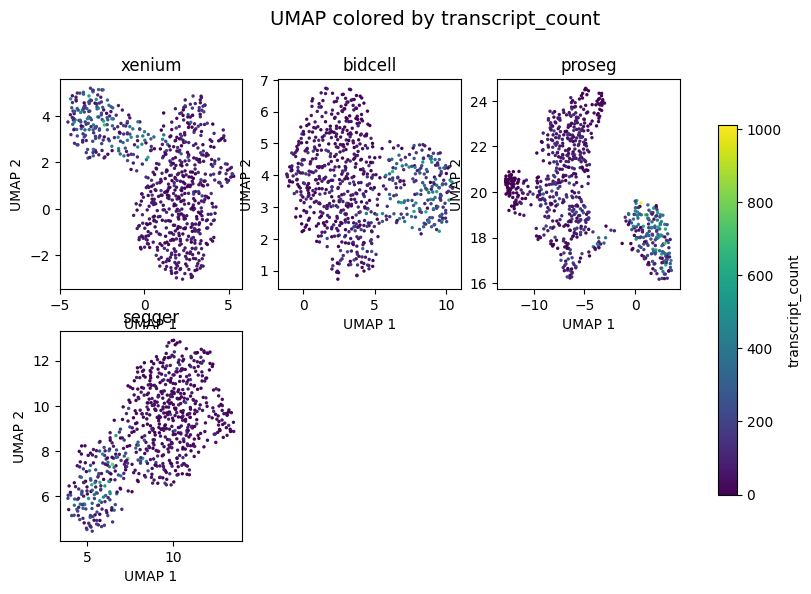
_ = segtraq.pl.umap(st_dict, color="transcript_count", legend=True, figsize=(10, 6))
[7]:
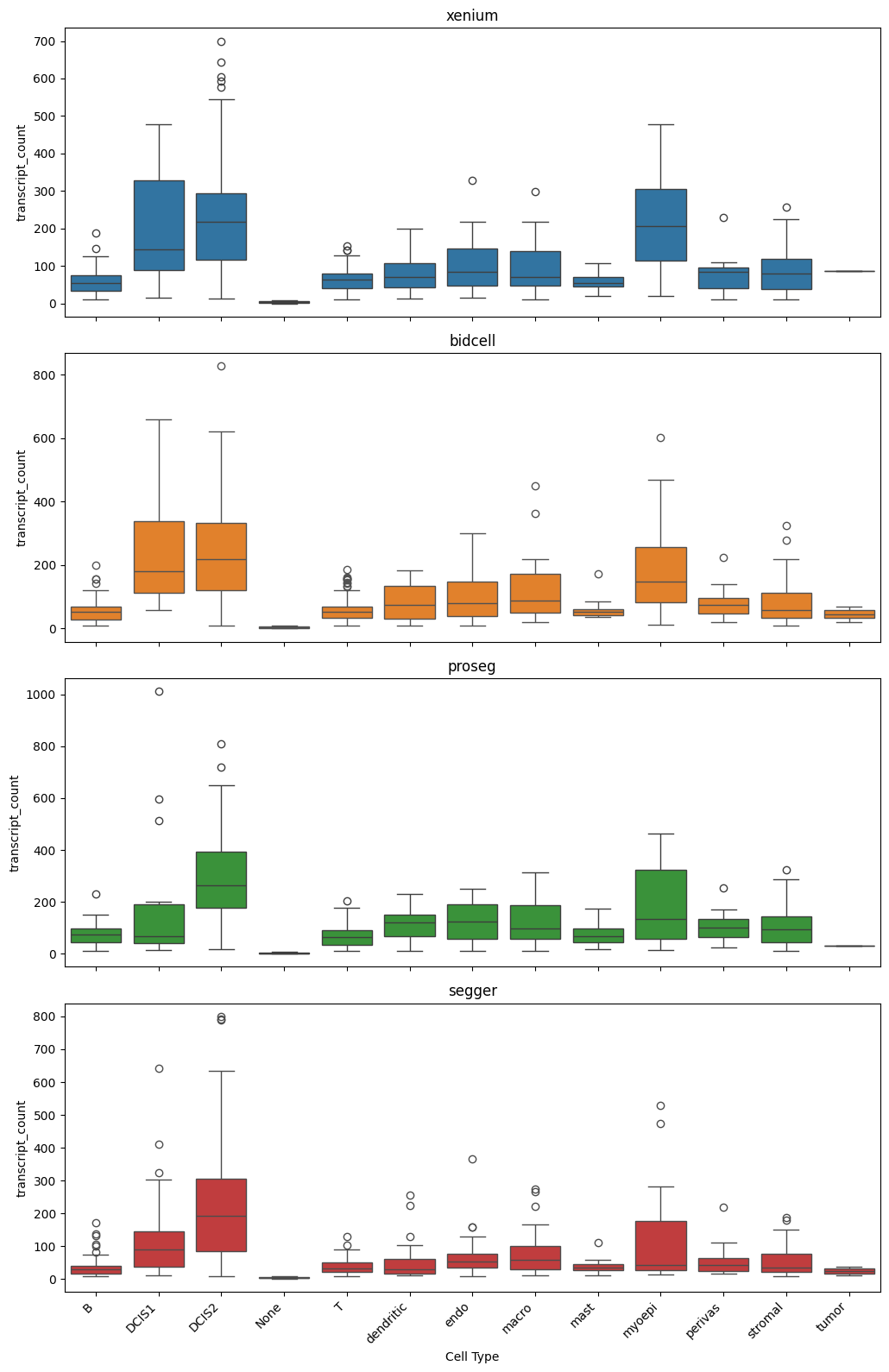
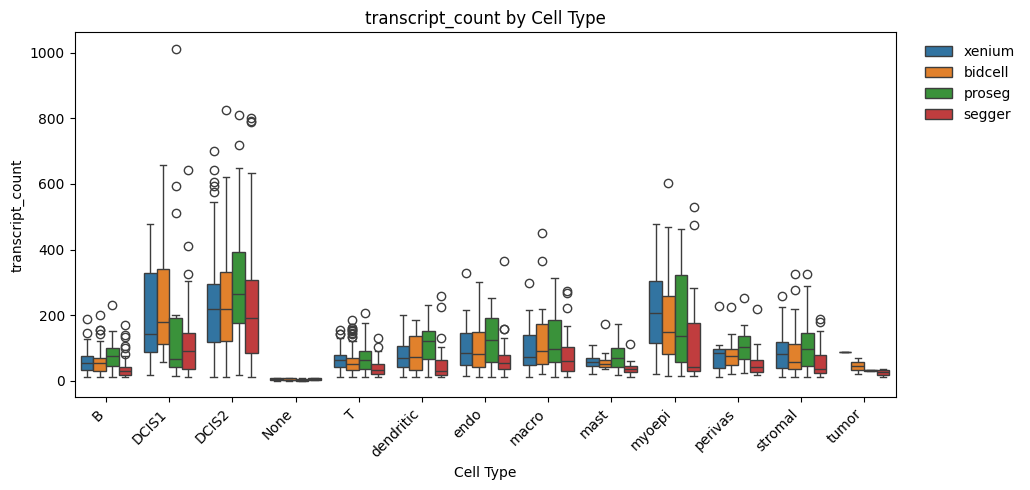
_ = segtraq.pl.boxplot(st_dict, celltype_col="transferred_cell_type", value_key="transcript_count")
[8]:
_ = segtraq.pl.boxplot_combined(st_dict, celltype_col="transferred_cell_type", value_key="transcript_count")
Session Info#
[9]:
print(sd.__version__) # spatialdata
0.7.2
