Module: Supervised Metrics#
Unlike scRNA-seq, spatial transcriptomics measurements are affected by the local tissue context. Transcripts can be captured outside their cell of origin due to transcript diffusion, 3D overlap, or segmentation inaccuracies, leading to mixed or contaminated expression profiles. As a result, spatial cells may partially resemble their neighbors.
The supervised (sp) module provides metrics to evaluate how well cell profiles in a spatial transcriptomics dataset agree with a reference single-cell RNA-seq (scRNA-seq) dataset with cell type annotations.
By comparing spatial expression profiles to a high-quality scRNA-seq reference, the supervised module aims to quantify this mismatch. Specifically, we use the scRNA-seq dataset as a clean reference, transfer cell type labels to the spatial data, and then compute metrics that measure:
how well each spatial cell matches its expected cell type,
how much its expression resembles other (neighboring) cell types, and
if it is possible to predict that a cell of one cell type is adjacent to a different cell type.
To follow along with this tutorial, you can download the data from here.
[1]:
%load_ext autoreload
%autoreload 2
[2]:
from pathlib import Path
import anndata as ad
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scanpy as sc
import seaborn as sns
import spatialdata as sd
import spatialdata_plot # noqa
import segtraq
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
Transfer labels from scRNA-seq to spatial transcriptomics#
We first load a spatial transcriptomics dataset (stored as a SpatialData object) and an annotated scRNA-seq reference dataset (stored as an AnnData object). For demonstration purposes, we subset the spatial dataset to a smaller region to reduce runtime.
Next, we initialize a SegTraQ object from the SpatialData. During initialization, the validate_spatialdata() method is automatically run to ensure that all required attributes (e.g. images_key, tables_key) are correctly specified.
We then transfer cell type labels from the scRNA-seq reference to the spatial dataset using run_label_transfer(). This method computes cell type–specific mean expression profiles in the reference and assigns each spatial cell to the cell type with the highest Pearson correlation to its expression profile.
Finally, we define a color scheme for visualizing the transferred cell type labels.
[3]:
# Load spatial transcriptomics dataset
sdata_ws = sd.read_zarr("../../data/xenium_5K_data/xenium.zarr/")
# Subset the dataset to a specific bounding box
bb_xmin = 800
bb_ymin = 1150
bb_w = 200
bb_h = 300
bb_xmax = bb_xmin + bb_w
bb_ymax = bb_ymin + bb_h
sdata = sdata_ws.query.bounding_box(
axes=["x", "y"],
min_coordinate=[bb_xmin, bb_ymin],
max_coordinate=[bb_xmax, bb_ymax],
target_coordinate_system="global",
)
# initialize SegTraQ object
st = segtraq.SegTraQ(sdata, images_key="image", tables_centroid_x_key="x_centroid", tables_centroid_y_key="y_centroid")
# Load scRNA-seq dataset
scRNAseq_data_path = Path("../../data/xenium_5K_data/BC_scRNAseq_Janesick.h5ad")
adata_ref = ad.read_h5ad(scRNAseq_data_path)
# Define color palette for cell types
col_celltype = {
"T": "#fb8072",
"B": "#bc80bd",
"macro": "#910290",
"dendritic": "#fdb462",
"mast": "#959059",
"perivas": "#fed9a6",
"endo": "#a6cee3",
"myoepi": "#2782bb",
"DCIS1": "#3c7761",
"DCIS2": "#66a61e",
"tumor": "#66c2a5",
"stromal": "#d45943",
"Unknown": "#808080",
}
no parent found for <ome_zarr.reader.Label object at 0x7fbcb9536510>: None
no parent found for <ome_zarr.reader.Label object at 0x7fbcb953bb10>: None
/home/meyerben/.local/share/uv/python/cpython-3.13.5-linux-x86_64-gnu/lib/python3.13/functools.py:934: UserWarning: The object has `points` element. Depending on the number of points, querying MAY suffer from performance issues. Please consider filtering the object before calling this function by calling the `subset()` method of `SpatialData`.
return dispatch(args[0].__class__)(*args, **kw)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/src/segtraq/utils.py:1255: UserWarning: Duplicate IDs detected in index 'cell_id' for shapes 'nucleus_boundaries'. Resetting and renaming index to `segtraq_id` to ensure uniqueness.
nucleus_shapes = _ensure_index(
sdata.tables[tables_key].obs["transferred_cell_type"].cell_type_key argument.tx_min, tx_max, gn_min, and gn_max.adata_ref.var_names andsdata.tables[tables_key].var_names. If they differ, specify the corresponding identifier columns using ref_ensemble_key and query_ensemble_key.None (default), var_names are used.[4]:
st.run_label_transfer(
adata_ref,
ref_cell_type="celltype_major", # Reference cell type column
ref_ensemble_key=None,
query_ensemble_key=None,
)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/src/segtraq/utils.py:383: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
ref_mean_df = counts_df.groupby("celltype").mean()
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/src/segtraq/SegTraQ.py:745: RuntimeWarning: Spatialdata table appears to contain raw counts. Counts will be log1p-transformed before running label transfer.Raw counts will be stored in `adata_q.layers["counts"]`.
result = _run_label_transfer(
Next, cell shapes can be plotted and colored by cell type based on a defined color palette. Some cells lack a transferred cell-type label (due to filtering low-count cells before label transfer); for compatibility with spatialdata-plot, we replace missing values (NaN) with “Unknown” before plotting.
[5]:
# Replace NaN with Unknown
s = st.sdata.tables["table"].obs["transferred_cell_type"]
if pd.api.types.is_categorical_dtype(s):
s = s.cat.add_categories(["Unknown"])
st.sdata.tables["table"].obs["transferred_cell_type_plot"] = s.fillna("Unknown")
labels = st.sdata.tables["table"].obs["transferred_cell_type_plot"].unique().astype(str).tolist()
cols = [col_celltype[lab] for lab in labels]
/tmp/ipykernel_27337/964490482.py:3: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(s):
[6]:
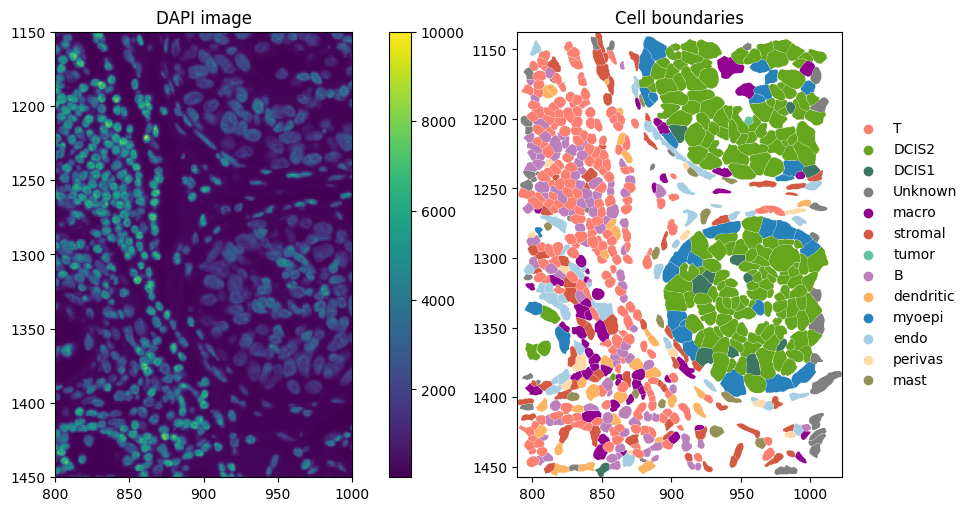
axes = plt.subplots(1, 2, figsize=(10, 5), constrained_layout=True)[1].flatten()
# Dapi image
st.sdata.pl.render_images("image").pl.show(ax=axes[0], title="DAPI image", coordinate_systems="global")
# Add link between table and spatial element
st.sdata.tables["table"].obs["region"] = "cell_boundaries"
st.sdata.set_table_annotates_spatialelement("table", region="cell_boundaries")
st.sdata.pl.render_shapes(
"cell_boundaries",
color="transferred_cell_type_plot",
palette=cols,
groups=labels,
outline_color="white", # outlines visible on black
outline_width=0.5,
).pl.show(ax=axes[1], title="Cell boundaries", coordinate_systems="global")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/spatialdata.py:477: UserWarning: Converting `region_key: region` to categorical dtype.
convert_region_column_to_categorical(table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata_plot/pl/utils.py:872: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = color_source_vector.map(color_mapping)
Spatial data are normalized and log transformed for some of the metrics below. The raw counts will be saved in sdata.tables[tables_key].layers["counts"].
[7]:
adata = st.sdata.tables[st.tables_key]
# Store raw counts
adata.layers["counts"] = adata.X.copy()
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
/tmp/ipykernel_27337/2280590036.py:4: UserWarning: Some cells have zero counts
sc.pp.normalize_total(adata, target_sum=1e4)
Computing cell type-specific markers#
Positive markers are genes that are consistently upregulated in one cell type relative to other cell types. To avoid composition bias arising from unequal cell type abundances, we perform pairwise comparisons between all cell types in the reference dataset. Marker detection can be performed using either differential expression (mode="de") or AUC-based scoring (mode="auc").
For each cell type, genes are selected by a voting scheme: a gene must be identified as upregulated in at least a fraction of pairwise contrasts (vote_fraction_pos, default = 0.5). To ensure that positive markers are representative of the cell type rather than driven by a small subset of cells, we additionally require that a gene is expressed (> 0) in at least min_pos_frac (default = 0.1) of cells of that type. To further increase specificity, we require that a positive marker is not
shared by more than t_pos × n_celltypes cell types. This yields markers that are both upregulated and cell-type–specific, without bias from cell type composition in the reference data.
Negative markers are derived from the same pairwise contrasts but are defined as genes that are effectively off in a focal cell type while being upregulated elsewhere. For each ordered pair of cell types (a, b), genes that are upregulated in a relative to b are considered negative-marker candidates for b if they are expressed (> 0) in at most max_neg_frac (default = 0.05) of cells of type b. To avoid ambiguous genes with inconsistent directionality, we further require
that a negative marker of b is not identified as upregulated in b relative to any other cell type across all ordered contrasts.
mode="auc", the minimum required AUC can be set via auc_pos_thresh (default = 0.9).mode="de", the differential expression method can be selected via method ("wilcoxon", "t-test", or "logreg"), with thresholds defined by logFC_pos_thresh (default = 1.0) and pval_adj_thresh (default = 0.05).All computations can be parallelized by specifying the number of jobs via n_jobs.
Only genes common to the scRNA-seq and spatial transcriptomics dataset will be considered to reduce compute time.
[8]:
# subsetting to commong genes
common_genes = adata.var_names[adata.var_names.isin(adata_ref.var_names)]
adata_ref = adata_ref[:, common_genes].copy()
[9]:
# get cell type-specific markers from the reference dataset
markers = segtraq.markers_from_reference(
adata_ref,
cell_type_key="celltype_major",
mode="de",
vote_fraction_pos=0.5,
min_pos_frac=0.1,
max_neg_frac=0.05,
n_jobs=16,
)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:531: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
A list of positive and negative markers computed for each cell type is shown below.
[10]:
for cell_type, sign in markers.items():
pos = sign["positive"]
neg = sign["negative"]
print(f"{cell_type} | positive: {', '.join(pos)}")
print(f"{cell_type} | negative: {', '.join(neg)}\n")
B | positive: MS4A1, BANK1, CD79A, TNFRSF13B, BLK, TNFRSF13C, POU2AF1, AIM2, PAX5, FCRL1, CD79B, FCRL2, P2RX5, POU2F2, PTPN6, CD19, FCRL5, SCIMP, CD22, FCMR, NIBAN3, CR1, KCNN4, CD27, SMCHD1, SPIB, DERL3, STAP1, FCRLA, NCOA3, COL4A3, FCRL3, ANKRD13A, CCDC141, ZBP1, PIM2, COL4A4, TNFRSF17, IRF4, CNR2, SYVN1, CD38, NCF1, CCR6, SLAMF6, CYSLTR1, COCH, CCDC50, BCL2, PDK1, TRAF5, CASP10, KCNA3, SLAMF1, TLR1, FCGR2B, GPR174, BTN3A1, CD47, SYNRG, APOBEC3C, NFKB2, CASP8, ICAM3, PTPN1, LPAR5, DMXL1, ORAI2
B | negative: AAR2, ABCA10, ABCA8, ABCB1, ABCC12, ABCC8, ABCC9, ABI3BP, ABTB2, AC007906.2, ACPP, ACVRL1, ADAM12, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRB1, ADGRE2, ADGRG6, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADRA2A, ADTRP, AHRR, AIF1, AKR1C3, ALDH1B1, ALDH7A1, ALPL, AMOT, ANGPT1, ANGPTL2, ANGPTL4, ANKRD6, ANXA3, ANXA8, APCDD1, APLNR, ARG2, ARL4D, ASPN, ATF3, ATL1, AUNIP, AVPR1A, AXIN2, BCL6B, BDKRB2, BLM, BMP2, BMPR1B, BNC2, BPIFB1, BRCA1, BRINP1, C1QTNF1, C3AR1, C4B, C5AR1, C7, CABP4, CABYR, CACNA1B, CACNA2D1, CACNG4, CADM3, CALB2, CALCRL, CALML3, CAMK1, CARD9, CAVIN2, CBS, CCDC136, CCL14, CCL19, CCL2, CCL22, CCL28, CCN3, CCN4, CCNE1, CCR4, CCR5, CD1E, CD209, CD274, CD302, CD33, CD40LG, CD7, CD8A, CDC25A, CDC6, CDH13, CDH23, CDH5, CDH6, CDK18, CDKL1, CDKN2B, CDKN3, CEBPA, CENPV, CEP112, CES1, CFC1, CHST8, CLCN5, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4A, CLEC4C, CLEC5A, CLEC7A, CLGN, CMA1, CMKLR1, CNN1, CNRIP1, CNTNAP1, COL10A1, COLQ, COMP, CPVL, CPXM2, CRB2, CREB5, CRYBG3, CSF2RA, CSF3R, CSPG4, CST7, CTLA4, CTSG, CTSW, CTTNBP2NL, CUBN, CUX2, CX3CL1, CX3CR1, CXCL2, CXCL9, CXCR6, CYP27A1, DCLK1, DDR2, DDX28, DHODH, DIO2, DLEC1, DLG4, DLK1, DLL4, DLX4, DMD, DNAH10, DNAH5, DNAH7, DNASE1L3, DNM1, DNMT3B, DOC2A, DPT, DPY19L2, DRAM1, DRC3, DSC3, DSG3, DTX1, DZIP1L, EBF2, ECRG4, EDN1, EDNRA, EFR3B, EGF, EGFL8, EGLN3, EGR2, ENPP3, EPB41L3, EPHA4, EPHB1, EREG, ERG, ERRFI1, ERVMER34-1, ESM1, ETV1, ETV5, F13A1, F2R, F2RL3, F7, FAM107A, FAM161A, FAP, FAT2, FAT4, FBLN2, FBXO16, FCER1A, FCGBP, FCGR3A, FGF1, FGF2, FGF7, FGFR4, FLT1, FLT3, FLT4, FMOD, FOLH1, FOLR2, FOXC1, FOXC2, FOXD2, FPR1, FREM1, FST, FZD1, GAB2, GABRD, GABRP, GADD45A, GADD45G, GHR, GIMAP8, GJC1, GLI2, GLS2, GNAI1, GNAO1, GPER1, GPR39, GPR4, GPRASP2, GPRC5B, GRAMD2B, GRIK4, GRM4, GRP, GUCY1A2, GXYLT2, GYG2, GZMA, GZMB, GZMK, HAS2, HAVCR2, HBEGF, HCK, HCN3, HDC, HEY1, HEYL, HGF, HLX, HMCN1, HMGN5, HNMT, HOXA5, HOXB4, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, HS6ST2, HSD11B2, HSPB8, ICOS, IDO1, IFITM1, IGF1, IKZF4, IL18, IL18R1, IL1R1, IL1RAP, IL1RL1, IL2RB, IL33, IL34, IL3RA, IL6R, INHBA, IRX1, ITGA2B, ITGA5, ITGA7, ITGB1BP2, ITIH5, ITPKA, JAM2, KCNC4, KCNF1, KCNH6, KCNJ8, KCNK10, KCNK15, KCNK17, KCNMA1, KCNMB1, KDR, KIAA0408, KIT, KITLG, KLHL13, KLK10, KLK6, KLRD1, KLRG1, LACC1, LAMB3, LAMP3, LATS2, LCN2, LCNL1, LDB3, LEF1, LGR4, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LOX, LPAR1, LPL, LRG1, LRRC15, LTBP1, LTK, LURAP1, LZTS1, MAFF, MAGI3, MALL, MAP1A, MAP2, MAP3K7CL, MAPK11, MAPK8IP1, MC1R, MECOM, MED12L, MELTF, MERTK, MET, MFAP5, MITF, MLXIPL, MMP11, MMP9, MMRN2, MNDA, MNS1, MPDZ, MPP1, MRC1, MRM3, MS4A2, MSR1, MSTN, MTNR1B, MUC16, NAV2, NBPF3, NCCRP1, NCF2, NECTIN3, NEDD4, NEIL3, NEK2, NEXN, NGFR, NLGN3, NLRP3, NMB, NOD2, NOS1AP, NOS3, NOSTRIN, NPNT, NPR1, NPY1R, NR1H3, NR4A3, NR5A2, NRN1, NRROS, NT5DC3, NTRK2, NXT2, OLFM1, OLFML3, OLR1, OPRD1, OSBP2, OSCP1, OSGIN1, OXTR, P2RX7, P2RY13, P2RY14, P2RY6, PALMD, PAMR1, PBK, PCDH10, PCDH17, PCDH18, PCDH8, PDE2A, PDE3A, PDGFRA, PDGFRL, PDZK1, PGAP1, PGBD1, PGF, PGR, PHEX, PHF21B, PHLPP1, PI16, PIEZO2, PILRA, PIMREG, PKP2, PLA2G7, PLAUR, PLD1, PLEKHH2, PLIN1, PLK4, PLVAP, POTEI, POTEJ, PPARG, PPFIA3, PPP1R1C, PPP2R3A, PRDM10, PRDM6, PRF1, PRICKLE1, PRKCQ, PRKD1, PRMT5, PROCR, PRODH, PROM1, PROS1, PROX1, PRRT2, PSRC1, PSTPIP2, PTAFR, PTCH1, PTGDR, PTGES, PTGFRN, PTGIR, PTGIS, PTK7, PTPRB, PTPRM, PYGL, RAD54L, RASD1, RCAN2, RECK, RELN, RFX2, RGS5, RIMS3, RNF180, RNF26, RNF39, ROBO4, ROR2, RPA3, RRAS, RTN1, RUNX1T1, SAC3D1, SAMD5, SATB2, SCGB1D2, SCN9A, SDHAF3, SELP, SEPTIN5, SERPINB5, SESN2, SFRP4, SH2D1A, SH2D2A, SH3RF2, SHANK3, SHD, SIGLEC1, SIGLEC9, SIM2, SIRPA, SKA3, SLAMF8, SLC11A1, SLC16A9, SLC18A2, SLC1A3, SLC26A2, SLC2A9, SLC35G2, SLC6A1, SLCO2A1, SLCO2B1, SLIT2, SLIT3, SMPD3, SOCS2, SOCS3, SOD3, SOSTDC1, SOX10, SOX17, SPAG1, SPAG17, SPATA18, SPOCK1, SPON1, SRPX, SSX2IP, STAC3, STEAP1, STMN3, STRIP2, STS, STX11, SUGCT, SULT1A1, SUV39H1, SVEP1, SYNCRIP, TAL1, TBC1D8B, TBX2, TBXA2R, TCL1A, TDO2, TEK, TEKT3, TEX30, TFAP2B, TFPI2, TFR2, TGFB2, TGFBR3, TGM2, TH, THBD, TIE1, TIGIT, TLE6, TLR2, TLR5, TLR9, TMEM119, TMEM200A, TMEM255B, TMEM98, TMPRSS13, TNFRSF10C, TNFSF13B, TNFSF15, TNFSF8, TP63, TPMT, TPPP3, TPSG1, TREM2, TRH, TRPC1, TRPC6, TSPAN18, TSPAN7, TTC23, TUBB2B, TWIST1, TWIST2, UBASH3A, UCN, UNC5A, VGLL3, VSIG4, VSNL1, VTCN1, WFDC1, WNT2, WNT4, ZFHX4, ZNF165, ZNF177, ZNF232, ZNF408, ZNF608, ZNF683
DCIS1 | positive: PRKAR1A, SLC16A1, MCM3AP, MUC16, SIAE, PSMD3, RMND1, DDX27, SCAF4, PCDH10, SCGB1D2, BMPR1B, EREG, YTHDF1, TIRAP, DLX4, OPRD1, POTEI, CACNA1D, GART, PPP1R9B, DPM1, WFS1, PER3, THYN1, ABCA10, BCAR3, DNAH7, SDHAF3, ROR2, VTCN1, HMGN5, PRODH, SMPD3, CACNG4, OSGIN1, FOXD2, KCNF1, PPFIA3, C4B, STAU1, EPHX1, RXRA, BCL3, P4HB, SRA1, SMARCA4, POR, PSENEN, DEPTOR, CANX, CAPN1, NT5DC2, MOB2, CYP27A1, TMEM209, PPP3CA, IFNGR2, TOLLIP, ADNP, CSE1L, PIH1D1, FAF2, IMPDH1, FAM53C, YIPF1, TCFL5, MAGI3, SPATA18, MC1R, TMPRSS13, ITGB1BP2, RAD54L, ARL4D, SERPINB5, RHOA, CTNNA1, MICAL2, SRC, COMT, PDIA3, IQGAP2, KIF5B, DIP2A, WWP1, TOP1, YWHAZ, RARA, CALM3, IRGQ, CCDC124, VEGFB, TM9SF4, DDX1, GOLPH3, CARM1, BCCIP, UBE2I, PRPF19, KCNK6, EIF3A, TMX2, STK35, PRRC2A, TCP1, ELMOD2, UBE2D2, DDB1, ALAD, URB1, BAD, IRF2BP1, ZHX3, LRRC59, WDR55, FN3KRP, UTP23, PRKCZ, DKC1, CCDC137, ABCB6, TBRG4, YIF1B, SIGMAR1, NOA1, CC2D1A, ELK1, RRM1, FOXRED1, LIG4, CDK7, SAC3D1, ERP27, KCNK15, LAMB3, DNAH5, MAPK8IP1, NOD2, LAMP1, GNA11, DBN1, STRN4, MAN1A1, POLR2E, DAG1, ATP1A1, TMEM109, UBR5, CSNK2B, AKT2, UPF1, SEPTIN9, CLCN3, NONO, ACVR1B, MRFAP1L1, FAF1, HYOU1, ITPR3, ABCD3, NCLN, GSK3A, UBA1, BSDC1, SLC37A4, HDAC2, XRCC1, GMDS, HDAC3, DDX21, AIMP2, SFXN1, PAIP2, TRUB1, PRKCI, HTATIP2, PIN1, AAR2, MGMT, DNAJC9, UBAP2, STK32C, GPR137, CKAP2, HGH1, MSH3, SNX15, CMSS1, PGBD1, TDO2, CDC25A, ZNF165, TLE6, TEX30, CYBC1, TCF3, SMARCC1, HSPA4, SYVN1, ILF3, MYBL2, DOK7, NOTCH3, PRKACA, WASL, EFNB2, FOXO3, RAB7A, NACC2, ECE1, TMEM123, TMEM106B, GSK3B, TSC2, SEC13, AP3D1, DVL1, MTA2, ELOC, NQO2, NUDCD2, FBL, EMD, RANGAP1, GTPBP4, PYCARD, ATP6V1H, MAEA, FBXW11, ACLY, CSNK2A1, KLF16, YIPF5, WDR12, JDP2, ERCC2, SLC39A8, SSX2IP, MIPOL1, RIPK2, DPAGT1, ZGRF1, POMT1, FANCI, RASD1, AMACR, PRDM10, RRAS, TBC1D8B, RNF26, NXT2, SUV39H1, TNFSF15, OSBP2, SATB2, CNN1, FAM161A, SPAG1, SHLD1, GTF2I, SNX8, EIPR1, PHF8, DDX31, ATPAF1, STK16, IPO5, OXSM, QPCTL, NSUN5, RBBP4, RHOH, GLCCI1, ZC3H12A
DCIS1 | negative: ABCA8, ABCB1, ABCC12, ABCC8, ABCC9, ABI3, ABI3BP, ABTB2, ACVRL1, ADA, ADAM12, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADCYAP1, ADGRB1, ADGRE2, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADRA2A, ADTRP, AIM2, AKR1C3, ALPL, ANGPT1, ANKRD6, ANXA8, APCDD1, APLNR, APOBEC3D, ASPN, ATL1, AVPR1A, BANK1, BCL11B, BCL2A1, BCL6B, BLK, BMP2, BMP6, BNC2, BPIFB1, BRINP1, C3AR1, C7, CALB2, CALCRL, CASP10, CAVIN2, CCDC136, CCDC141, CCL14, CCL19, CCL2, CCN3, CCN4, CCR1, CCR4, CCR5, CD19, CD1C, CD1D, CD1E, CD209, CD22, CD226, CD247, CD27, CD274, CD28, CD302, CD33, CD38, CD3G, CD40LG, CD5, CD7, CD72, CD79A, CD79B, CD86, CDH13, CDH23, CDH5, CDH6, CDK18, CDKL1, CENPV, CEP112, CHST8, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4C, CLEC5A, CLEC7A, CMA1, CMKLR1, CNR2, CNRIP1, CNTNAP1, COCH, COL10A1, COL4A3, COL4A4, CPVL, CPXM2, CR1, CRB2, CREB5, CSF2RA, CST7, CTLA4, CTSG, CTSW, CUBN, CUX2, CX3CR1, CXCL2, CXCL9, CXCR3, CXCR6, CYSLTR1, DCLK1, DERL3, DIO2, DLEC1, DLK1, DLL4, DNAH10, DNASE1L3, DPT, DPY19L2, DTX1, EBF2, EDN1, EDNRA, EGF, EGR2, ENOX1, ENPP3, EPB41L3, EPHB1, ERG, ESM1, F2RL3, FAM107A, FAT4, FCER1A, FCGR2B, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGF2, FGF7, FGR, FLI1, FLT1, FLT3, FLT4, FOLH1, FOLR2, FOXC2, FPR1, FREM1, FUT7, GAB2, GABRD, GBP5, GHR, GIMAP8, GJC1, GLI2, GPR174, GPR34, GPR4, GRIK4, GUCY1A2, GYG2, GZMA, GZMB, GZMK, HAS2, HBEGF, HCK, HDC, HEYL, HGF, HHEX, HLX, HMCN1, HOXA5, HOXB4, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, HSPA6, HSPB8, ICAM2, ICOS, IDO1, IFITM1, IGF1, IL12RB1, IL18R1, IL1RL1, IL21R, IL2RB, IL33, IL3RA, IL6R, INHBA, IRF4, IRF5, ITGA7, ITIH5, ITK, ITPKA, JAM2, KCNA3, KCNJ8, KCNK10, KCNK17, KCNN3, KDR, KIAA0408, KIT, KLK10, KLRD1, KLRG1, LAIR1, LAMP3, LCK, LCN2, LCNL1, LDB3, LEF1, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LOX, LPAR1, LPL, LRRC15, LTBP1, LTK, LURAP1, LY96, LZTS1, MAP1A, MAP2, MAP3K7CL, MED12L, MELTF, MERTK, MFAP5, MLXIPL, MMP9, MNDA, MPP1, MRC1, MS4A1, MS4A2, MTNR1B, NCCRP1, NCF1, NCF2, NFATC1, NIBAN3, NLGN3, NLRP3, NOS3, NPR1, NR4A3, NR5A2, NRN1, NRROS, OLR1, P2RX1, P2RX5, P2RX7, P2RY13, P2RY14, PAX5, PCDH17, PCDH18, PCDH8, PDE2A, PDE3A, PGF, PGR, PHEX, PI16, PIEZO2, PILRA, PKP2, POTEJ, POU2AF1, POU2F2, PRDM6, PRF1, PRICKLE1, PRKCQ, PRKD1, PROCR, PROX1, PTCH1, PTGDR, PTGIR, PTGIS, PTGS1, PTPRB, RCAN2, RECK, RELN, RGS5, RIMS3, RIPK3, ROBO4, RUNX1T1, S1PR1, SCIMP, SCN9A, SDK2, SELP, SEMA7A, SFRP4, SH2D1A, SH2D2A, SHANK3, SHD, SIGLEC1, SIGLEC6, SIGLEC9, SIM2, SLAMF1, SLAMF6, SLAMF8, SLC11A1, SLC18A2, SLC1A3, SLC2A3, SLC35G2, SLC6A1, SLC7A7, SLCO2A1, SLIT3, SOCS2, SOD3, SOSTDC1, SOX17, SOX5, SPIB, SPOCK1, SPON1, SRPX, STAP1, STAT5A, STRIP2, STX11, SUGCT, TAL1, TBX2, TBXA2R, TCL1A, TEK, TFAP2B, TFR2, TGFB2, TGFBR3, TGM2, TH, THBD, THEMIS, TIE1, TIGIT, TLR1, TLR7, TLR9, TMEM119, TMEM200A, TMEM255B, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFSF8, TPSG1, TREML2, TRH, TRPC1, TRPC6, TSPAN18, TSPAN7, TUBB2B, TWIST1, TWIST2, UBASH3A, VGLL3, VSIG4, VSNL1, WFDC1, WNT2, WNT4, WNT5B, ZBP1, ZFHX4, ZNF683
DCIS2 | positive: NAT1, TRIB3, LRG1, PDZK1, NPY1R, BPIFB1, PBX1, TBX3, RAB11FIP1, API5, IKZF2, FIBCD1, NAT10, FUT8, SOX2, AXIN2, CACNA1B, AURKA, KCNK15, NOS1AP, EGF, HS6ST2, SIM2, ANXA3, FGFR4, GPER1, NEK2, F7, GPR39, HCN2, SPAG17, TRH, CORO1B, MYO1C, FADD, IARS2, REPS1, GSDMB, SMC4, GSR, ZNF281, NSD2, TFPI2, GALNT4, ABTB2, GADD45G, CIP2A, PTGES, EFR3B, UCN, PDGFRL, CDCA2, UNC5A, PLIN1, CBS, CCL22, STAC3, OSGIN1, KIAA0408, CFC1, PLK4, SLC16A9, MLXIPL, KCNH6, CTTN, CD46, KDM5B, ADIPOR1, VAPB, CKAP5, CCDC124, HSPD1, PSMD4, GFPT1, MTCH2, EEF1AKNMT, SLC25A1, UFC1, TMEM101, SF3B4, CDK8, ATG16L1, YWHAZ, HBS1L, PSEN2, C1QBP, RBCK1, CNIH4, CBX5, DNPEP, EPRS, ALAD, ADIPOR2, ECM1, STMN1, MSH6, LSM12, SNAP47, THUMPD3, PBDC1, DVL2, TICAM1, RRS1, UBE2T, GCNT2, TRPV1, CENPF, PRRT2, GSTZ1, SPAG5, GHR, STS, CABP4, HSD11B2, AHRR, AURKB, PBK, NMB, CDKN3, BDKRB2, WNT4, DOC2A, DRC3, ACPP, MSTN, EIF2AK3, RHOA, PRKAR1A, CTNNA1, HIPK3, WWP1, ULK1, STAU1, CDK4, MAP4K3, SSRP1, CKAP4, KXD1, GOLPH3, DNAJA1, PSMB5, GLUD1, CASK, TOP1, SH3BP5L, PSENEN, POLDIP2, IDH1, SHARPIN, FDPS, KDM6B, APTX, KRAS, CDK16, KRTCAP2, CBX3, BRI3BP, ZNF687, GLA, NTPCR, USP10, NRAS, FECH, CCT2, RITA1, MID2, GPN1, MCM6, SHMT1, REEP4, WDHD1, MAGI3, OLFM1, RIPK3, HCN3, ZNF408, ZNF232, NOSTRIN, ADRA2A, PRDM6, CD82, LPCAT1, DOK7, ANXA2, TJP1, PON2, RAC1, EPHX1, SQSTM1, CRK, PLEKHB2, NRIP1, WASL, IRGQ, TOM1L2, RAB7A, PGD, TMUB2, BAG3, RAB27A, MYLIP, CAT, CLIC4, EIF2S1, SP1, SNX30, PIK3CB, DHX9, SLC40A1, SRI, EIF2AK2, GSK3B, HTT, SLC39A9, TPCN1, NBL1, EIF3A, NQO2, DACH1, TRAFD1, POLE, SLMAP, PFKP, FH, NAMPT, HEXIM1, TRIM28, FXR2, PYCARD, DHX34, MINDY2, RUVBL1, TNFRSF6B, SLC20A2, LRRC59, TSPYL5, KCNK6, RRP15, HPRT1, HAX1, DDIT3, NUDT1, GMFB, TMEM120B, RIPK2, RAD18, NEDD4, TMEM97, FAM98A, YJEFN3, CDK7, RFC5, BCL2L12, LTBP1, PPP2R3A, TPMT, LGR4, SULT1A1, OSCP1, CDKN2B, ARG2, FBXO16, MED12L, RBM38, CYP24A1, HHEX, ANKS6, NT5E, MTNR1B, PPP1R15B, MCM7, SNX8, FDFT1, GMDS, ATPSCKMT, SHMT2, BLM, ERP27
DCIS2 | negative: ABCA10, ABCA8, ABCC9, ABI3, ABI3BP, AC007906.2, ACSL4, ACVRL1, ADA, ADAM12, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRE2, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADTRP, AIM2, AKR1C3, AKT3, ALPL, ANGPT1, ANGPTL2, ANKRD6, ANXA8, APLNR, ASPN, ATL1, AVPR1A, BANK1, BCL11B, BCL2A1, BCL6B, BLK, BMP2, BMP6, BNC2, BRINP1, C1QTNF1, C3AR1, C7, CADM3, CALB2, CALCRL, CARD9, CAVIN2, CCDC136, CCDC141, CCL14, CCL19, CCL2, CCN3, CCN4, CCR1, CCR4, CCR5, CD19, CD1C, CD1D, CD1E, CD209, CD226, CD247, CD27, CD274, CD28, CD302, CD33, CD38, CD3D, CD3G, CD40LG, CD5, CD72, CD79A, CD79B, CD86, CD8A, CDH13, CDH23, CDH5, CDH6, CDKL1, CEP112, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4C, CLEC5A, CLEC7A, CMA1, CMKLR1, CNR2, CNRIP1, CNTNAP1, COCH, COL10A1, COL4A3, COL4A4, CPVL, CPXM2, CR1, CRB2, CRYBG3, CSF2RA, CST7, CTLA4, CTSG, CTSW, CUBN, CUX2, CX3CL1, CX3CR1, CXCL2, CXCL9, CXCR3, CXCR6, CYSLTR1, DCLK1, DERL3, DIO2, DLK1, DLX4, DMD, DNASE1L3, DPT, DPY19L2, DSG3, DTX1, EBF2, ECRG4, EDNRA, EGR2, ENOX1, ENPP3, EPHB1, ESM1, ETV1, ETV5, F2RL3, FAM107A, FAP, FAT4, FCER1A, FCGR2B, FCGR3A, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGF2, FGF7, FGR, FLI1, FLT1, FLT3LG, FLT4, FOLH1, FOLR2, FOXC2, FPR1, FREM1, FUT7, GABRD, GBP5, GIMAP8, GJC1, GLI2, GPR174, GPR34, GPR4, GRP, GUCY1A2, GZMA, GZMB, GZMK, HAS2, HBEGF, HCK, HDC, HEY1, HEYL, HGF, HLX, HMCN1, HOXA5, HOXB4, HOXD10, HOXD9, HPGDS, HPSE, HPSE2, HS3ST3A1, ICAM2, IDO1, IGF1, IL12RB1, IL18, IL18R1, IL1RL1, IL21R, IL2RB, IL33, IL34, IL3RA, IL6R, INHBA, IRF4, IRF5, ITGA7, ITIH5, ITK, JAM2, KCNA3, KCNJ8, KCNK10, KCNK17, KCNMB1, KCNN3, KDR, KIT, KLK10, KLK6, KLRD1, KLRG1, KLRK1, LACC1, LAIR1, LAMP3, LCK, LCN2, LCNL1, LDB3, LEF1, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LPAR1, LPAR5, LPL, LRRC15, LTK, LURAP1, LY96, LZTS1, MALL, MAP1A, MAP3K7CL, MERTK, MFAP5, MMP9, MMRN2, MNDA, MPP1, MRC1, MS4A1, MS4A2, MUC16, NCCRP1, NCF1, NEXN, NFATC1, NIBAN3, NLRP3, NOD2, NOS3, NPNT, NPR1, NR4A3, NR5A2, NRN1, NRROS, NTRK2, OLR1, OPRD1, P2RX1, P2RX5, P2RX7, P2RY13, P2RY14, P2RY6, PALMD, PAMR1, PAX5, PCDH10, PCDH17, PCDH18, PDE2A, PDE3A, PDGFRA, PGF, PGR, PHEX, PI16, PIEZO2, PLEKHH2, POTEJ, POU2AF1, POU2F2, PPP1R1C, PRF1, PRICKLE1, PRKCQ, PROCR, PROS1, PROX1, PTCH1, PTGDR, PTGIR, PTGIS, PTGS1, PTPRB, RCAN2, RECK, RELN, RGS5, RIMS3, RNF180, ROBO4, ROR2, RTN1, RUNX1T1, S1PR1, SAMD5, SATB1, SCGB1D2, SCIMP, SCN9A, SDHAF3, SDK2, SELP, SEMA7A, SFRP4, SH2D1A, SHANK3, SHD, SIGLEC1, SIGLEC6, SIGLEC9, SLAMF1, SLAMF6, SLAMF8, SLC11A1, SLC18A2, SLC1A3, SLC2A3, SLC35G2, SLC6A1, SLC7A7, SLCO2A1, SLIT2, SMPD3, SOD3, SOSTDC1, SOX10, SOX17, SPIB, SPOCK1, SPON1, SRPX, STAP1, STAT4, STMN3, STX11, SUGCT, SVEP1, TAL1, TBX2, TBXA2R, TCL1A, TDO2, TEK, TFAP2B, TGFB2, TGFBR3, TH, THBD, THEMIS, TIE1, TIGIT, TLR1, TLR7, TLR9, TMEM119, TMEM200A, TMEM255B, TMEM98, TNFRSF13B, TNFRSF17, TNFSF8, TPPP3, TPSG1, TRBC1, TREM2, TREML2, TRPC1, TRPC6, TSPAN18, TSPAN7, TUBB2B, TWIST1, TWIST2, UBASH3A, VGLL3, VSIG4, VSNL1, WFDC1, WNT2, WNT5B, ZBP1, ZFHX4, ZNF608, ZNF683
T | positive: IL7R, CD2, TRAC, CD96, CD3E, ZAP70, CD3D, CD6, TRBC1, KLRK1, THEMIS, STAT4, ITK, LCK, TCF7, CD3G, CD28, BCL11B, CD5, IL2RB, GZMA, CD247, CD8A, RBL2, GBP5, RASGRP1, CASP8, PRDM1, SLAMF1, SLFN5, LAT, FYN, TRAF1, LEPROTL1, AOAH, CD40LG, CD226, KLRD1, CD7, ZNF683, CCR4, CTSW, GZMK, KLRG1, TIGIT, PRKCQ, CXCR6, PRF1, SH2D1A, PBXIP1, SH2D2A, FLT3LG, UBASH3A, GPR174, KCNA3, FCMR, SIGIRR, CCR5, NFATC2, CST7, CDC14A, CTLA4, TNFSF8, PTGDR, CXCR3, RUNX3, APOBEC3G, SYNRG, SLAMF6, CD27, BTN3A1, SEMA4D, SMCHD1, ZBP1, ADGRE5, SATB1, ICOS, CCND2, LEF1, IL12RB1, KLF12, ZBTB25, PLCG1, CASK, DOCK9, S1PR1, IL21R, PDCD4, APOBEC3D, CD58, OGT, TMEM173, STAT5B, CD47, ERN1, ICAM2, MBNL1, IKBKE, CYLD, TAP1, ICAM3
T | negative: AAR2, ABCA10, ABCA8, ABCB6, ABCC12, ABCC8, ABCC9, ABI3BP, ABL2, ABTB2, AC007906.2, ACKR3, ACP5, ACPP, ACVR1B, ACVRL1, ADAM12, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRB1, ADGRB2, ADGRE2, ADGRG6, ADGRL4, ADIPOR2, ADORA2B, ADORA3, ADRA1B, ADRA2A, ADTRP, AGAP1, AHRR, AIF1, AIM2, AIMP2, AKR1C3, ALDH1B1, ALDH2, ALDH7A1, ALPK1, ALPL, AMACR, AMOT, ANGPT1, ANGPTL2, ANGPTL4, ANKRD6, ANKS6, ANXA3, ANXA8, AP1S3, APCDD1, APEX2, APLNR, ARG2, ARL4D, ASPN, ATAD3A, ATF3, ATL1, ATPAF1, AUNIP, AURKA, AURKB, AVPR1A, AXIN2, BAD, BAG3, BANK1, BARD1, BCAR1, BCAR3, BCL2A1, BCL2L12, BCL6B, BDKRB2, BLK, BLM, BLOC1S2, BMP2, BMP6, BMPR1B, BNC2, BPIFB1, BRCA1, BRCA2, BRINP1, BTRC, C1QC, C1QTNF1, C3AR1, C4B, C5AR1, C7, CABP4, CABYR, CACNA1B, CACNA1D, CACNA2D1, CACNG4, CADM3, CALB2, CALCRL, CALML3, CAMK1, CAMSAP2, CARD9, CAVIN2, CBS, CCDC137, CCDC170, CCDC50, CCDC80, CCL14, CCL19, CCL2, CCL22, CCL28, CCN3, CCN4, CCNE1, CCR1, CCT2, CD14, CD163, CD19, CD1C, CD1D, CD1E, CD209, CD22, CD274, CD302, CD33, CD38, CD72, CD79A, CD79B, CD86, CDAN1, CDC25A, CDC6, CDCA2, CDCA7L, CDH13, CDH23, CDH5, CDH6, CDK18, CDK7, CDK8, CDKN1A, CDKN2B, CDKN3, CEBPA, CEMIP2, CENPF, CENPV, CEP112, CEP170, CES1, CFAP20, CFC1, CHST8, CIP2A, CKAP2, CKAP4, CLCN5, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4A, CLEC4C, CLEC5A, CLEC7A, CLGN, CLIC4, CMA1, CMKLR1, CMSS1, CNIH4, CNN1, CNR2, CNRIP1, CNTNAP1, COBLL1, COCH, COL10A1, COL4A3, COL4A4, COLQ, COMP, CORO1C, CPVL, CPXM2, CR1, CRB2, CREB5, CRYBG3, CSE1L, CSF1R, CSF2RA, CSF3R, CSPG4, CTDSPL, CTNNAL1, CTSG, CTTNBP2NL, CUX2, CX3CL1, CX3CR1, CXCL2, CXCL9, CYCS, CYP1B1, CYP24A1, CYP27A1, CYSLTR1, DACH1, DAG1, DBN1, DCLK1, DDIT3, DDR2, DDX28, DEPTOR, DERL3, DHODH, DHX57, DIO2, DLEC1, DLG4, DLK1, DLL4, DLX4, DMD, DMXL2, DNAH10, DNAH5, DNAH7, DNAJB4, DNASE1L3, DNM1, DNMT3B, DOC2A, DOK7, DPAGT1, DPT, DPY19L2, DRAM1, DRC3, DSC3, DSG3, DTX1, DUSP5, DUSP6, DVL2, DZIP1L, EBF2, ECRG4, EDN1, EDNRA, EFNB2, EFR3B, EGF, EGFL8, EGLN3, EGR2, EIF2AK3, ELK1, ENOX1, ENPP3, EPAS1, EPB41L3, EPHA4, EPHB1, EPHX1, ERCC2, EREG, ERG, ERLIN2, ERRFI1, ERVMER34-1, ESM1, ETV1, ETV5, F13A1, F2RL3, F7, FAM107A, FAM117B, FAM161A, FAM53C, FAM98A, FANCC, FANCD2, FANCI, FAP, FAT2, FAT4, FBLN2, FBXO16, FBXO25, FBXW11, FCER1A, FCGR2A, FCGR2B, FCGR3A, FCRL1, FCRL2, FCRL5, FCRLA, FECH, FGF1, FGF2, FGF7, FGFR4, FGR, FH, FHL2, FIBCD1, FIBP, FLCN, FLT1, FLT3, FLT4, FMO5, FMOD, FOLH1, FOLR2, FOXC1, FOXC2, FOXD2, FOXRED1, FPR1, FREM1, FRMD6, FST, FUCA1, FUT7, FZD1, GAB2, GABRD, GABRP, GADD45A, GADD45G, GCNT2, GHR, GJC1, GLA, GLI2, GLOD4, GLS2, GMFB, GNA11, GNAI1, GNAO1, GPER1, GPN1, GPR137, GPR34, GPR39, GPR4, GPRASP2, GPRC5B, GRAMD2B, GRIK4, GRM4, GRP, GSTZ1, GUCY1A2, GXYLT2, GYG2, HAS2, HAVCR2, HBEGF, HCK, HCN2, HCN3, HDAC2, HDC, HEY1, HEYL, HGF, HGH1, HHEX, HLX, HMCN1, HMGA1, HMGN5, HMOX1, HNMT, HOXA5, HOXB4, HOXD10, HOXD9, HPGD, HPGDS, HPRT1, HPSE, HPSE2, HS3ST3A1, HS6ST2, HSD11B2, HSPA6, HSPB8, HTATIP2, IDE, IDH1, IDO1, IFNGR2, IGF1, IKZF4, IL18, IL1R1, IL1RAP, IL1RL1, IL33, IL34, IL3RA, IMPDH1, INHBA, IPO5, IRF2BP1, IRF4, IRF5, IRX1, ITGA2B, ITGA7, ITGB1BP2, ITGB5, ITIH5, ITPKA, JAM2, JDP2, KCNAB3, KCNF1, KCNH6, KCNJ8, KCNK10, KCNK15, KCNK17, KCNK6, KCNMA1, KCNMB1, KCNN3, KDR, KIAA0408, KIT, KITLG, KLF16, KLF4, KLHL13, KLK10, KLK6, KMO, LACC1, LAMB3, LAMP3, LANCL2, LATS2, LCAT, LCN2, LCNL1, LDB3, LGALS9, LGMN, LGR4, LIF, LIFR, LIG4, LILRA4, LILRB2, LILRB4, LILRB5, LIPE, LMO2, LOX, LOXL1, LPAR1, LPL, LRG1, LRP6, LRRC15, LRRC59, LSM12, LTBP1, LTBP2, LTK, LURAP1, LY96, LZTS1, MAFB, MAFF, MAGI3, MALL, MANEA, MAP1A, MAP2, MAP3K20, MAP3K7CL, MAP4K3, MAPK11, MAPK12, MAPK8IP1, MC1R, MECOM, MED12L, MEIS1, MELTF, MERTK, MET, METTL18, MFAP5, MGMT, MID2, MIPOL1, MITF, MLF1, MLXIPL, MMP11, MMP9, MMRN2, MNDA, MNS1, MPDU1, MPDZ, MPP1, MRC1, MRM3, MRTFB, MS4A1, MS4A2, MS4A6A, MSH3, MSR1, MSTN, MTCH2, MTNR1B, MUC16, MYBL2, MYOF, MZF1, NACC2, NAIF1, NAIP, NAMPT, NAT1, NAV2, NBPF3, NCCRP1, NCF1, NCF2, NDRG2, NECTIN3, NEDD4, NEIL3, NEK2, NEK8, NEXN, NGFR, NIBAN3, NIFK, NLGN3, NLRP3, NMB, NOA1, NOD1, NOD2, NOS1AP, NOS3, NOSTRIN, NOTCH3, NPNT, NPR1, NPY1R, NR1H3, NR4A3, NR5A2, NRIP1, NRN1, NRROS, NSUN5, NT5DC2, NT5DC3, NT5E, NTPCR, NTRK2, NUDT1, NXT2, OCRL, OLFM1, OLFML3, OLR1, OPRD1, OSBP2, OSCP1, OSGIN1, OXSM, OXTR, P2RX1, P2RX7, P2RY13, P2RY14, P2RY6, PALMD, PAMR1, PAX5, PBDC1, PBK, PCDH10, PCDH17, PCDH18, PCDH8, PDE2A, PDE3A, PDGFB, PDGFRA, PDGFRL, PDZK1, PER3, PGAP1, PGBD1, PGD, PGF, PGM1, PGR, PHB2, PHEX, PHF21B, PHLPP1, PI16, PIEZO2, PILRA, PIMREG, PINK1, PKP2, PLA2G7, PLAUR, PLD1, PLEKHA1, PLEKHH2, PLIN1, PLK4, PLVAP, PNPT1, POLD1, POMT1, POTEI, POTEJ, POU2AF1, PPARG, PPFIA3, PPM1H, PPP1R1C, PPP2R3A, PRDM10, PRDM6, PRICKLE1, PRKCI, PRKD1, PRMT5, PROCR, PRODH, PROM1, PROS1, PROX1, PRPS2, PRRT2, PSEN2, PSENEN, PSRC1, PSTPIP2, PTAFR, PTCH1, PTGES, PTGFRN, PTGIR, PTGIS, PTGS1, PTK2, PTK7, PTPN11, PTPRB, PTPRM, PVR, PYCARD, PYGL, QPCTL, R3HDM1, RAD18, RAD54L, RAPGEF3, RASD1, RBBP4, RBBP5, RCAN2, RCN2, RCOR1, RECK, REEP4, RELN, RFC4, RFC5, RFX2, RGS5, RIMS3, RIPK2, RITA1, RMND1, RNF180, RNF26, RNF39, ROBO4, ROR2, RPA3, RRAS, RRP15, RRS1, RTN1, RUBCN, RUNX1T1, RWDD4, SAC3D1, SAMD5, SATB2, SCGB1D2, SCIMP, SCN9A, SDHAF3, SDK2, SELP, SEMA7A, SEPTIN5, SERPINB5, SESN2, SFRP4, SH3RF2, SHANK3, SHD, SHMT1, SHMT2, SIAE, SIGLEC1, SIGLEC6, SIGLEC9, SIGMAR1, SIM2, SIRPA, SIRT3, SKA3, SLAMF8, SLC11A1, SLC16A1, SLC16A9, SLC18A2, SLC1A3, SLC20A2, SLC29A1, SLC2A9, SLC35G2, SLC37A4, SLC39A11, SLC40A1, SLC43A2, SLC6A1, SLC7A7, SLCO2A1, SLCO2B1, SLIT2, SLIT3, SMPD3, SNX10, SNX15, SNX8, SOCS2, SOCS3, SOD3, SOSTDC1, SOX10, SOX17, SOX2, SOX5, SPAG1, SPAG17, SPAG5, SPATA18, SPATS2L, SPECC1, SPIB, SPOCK1, SPON1, SRC, SRPX, SSBP3, SSX2IP, STAC3, STAP1, STEAP1, STK16, STK32C, STMN1, STRIP2, STS, STX11, SUGCT, SULT1A1, SUV39H1, SVEP1, SYNCRIP, TAL1, TBC1D8B, TBX2, TBX3, TBXA2R, TBXAS1, TCF7L2, TCFL5, TCL1A, TDO2, TEK, TEKT3, TEX30, TFAM, TFAP2B, TFPI2, TFR2, TGFB2, TGM2, TH, THBD, THBS2, TICAM1, TIE1, TIRAP, TJP1, TLE6, TLR1, TLR2, TLR5, TLR7, TLR9, TMEM101, TMEM119, TMEM120B, TMEM192, TMEM255B, TMEM97, TMEM98, TMPRSS13, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFRSF21, TNFRSF6B, TNFSF13B, TNFSF15, TOLLIP, TOR3A, TP63, TPCN1, TPMT, TPPP3, TPSG1, TRAF3IP2, TREM2, TREML2, TRH, TRIB3, TRIM28, TRPC1, TRPC6, TRPM2, TRPV1, TRUB1, TSPAN18, TSPAN7, TSPAN9, TSPYL5, TTC23, TUBB2B, TUBG1, TWIST1, TWIST2, UBAP2, UBE2T, UCN, UMPS, UNC5A, VAMP5, VDR, VGLL3, VSIG4, VSNL1, VTCN1, WDHD1, WDR12, WDR81, WFDC1, WFS1, WNT2, WNT4, WNT5B, YAP1, YES1, YIPF1, YJEFN3, YTHDF1, ZBTB10, ZFAT, ZFHX3, ZFHX4, ZFYVE26, ZGRF1, ZHX3, ZNF165, ZNF177, ZNF232, ZNF408, ZNF608, ZSWIM6
dendritic | positive: CSF2RA, CLEC4C, GZMB, IRF7, IL3RA, LILRA4, RUNX2, ZDHHC17, FLT3, CCDC50, FUT7, ZFAT, MAPKAPK2, CLCN5, ERN1, TNFRSF21, SCN9A, SHD, SPIB, NPC1, EPHB1, LCNL1, DNASE1L3, P2RY14, DUSP5, NIBAN3, CXCR3, ETV6, TLR7, RASD1, LAMP3, MAP3K14, PHEX, MRTFA, IFNAR2, KCNK17, SLC7A5, MDFIC, BTAF1, TFRC, PTPN1, IRF4, TCL1A, DERL3, MALT1, MX1, SMPD3, ZFYVE26, IDO1, MAP1A, CEP128, LTK, BCL2L11, TLR9, RELB, RUBCN, ADA, MS4A6A, VEGFB, ALDH2, LAIR1, CTSC, CMKLR1, TRAF1, JAK2, CORO1C, MAN2B1, KIF2A, UCP2, IFI44L, N4BP2, PI4KA, IL7R, PLEKHO1, IL6R, HS3ST3A1, NR4A3, CUX2, KCNK10, COBLL1, RIMS3, SEMA7A, NIN, ZMIZ2, BLK, NCOA3, TYMP, CNP, ZBTB33, ZBTB10, NFKB1, LILRB4, FCER1A, OGT, IRF1, P2RX1, ABCA7, ABCC4, BIRC2, WDR81, TNFSF13B, CDC14A, EIF4ENIF1, HCK, NCCRP1, CD55, RFX5, CYSLTR1, NCF1, TNFRSF17, ITCH, CAT, ULK1, DNM2, TYK2, NDRG1, CDKN1A, P2RY6, CCL19, PTGIR, HLX, CD274, IL21R, PLEKHA2, WNT5B, BCL2A1, TREML2, SNX29, PIM2, PPP1R9B, TAP1, CDCA7L, PFKFB3, IRF2BPL, NR3C1, HMGN3, ERCC1, TAB2, TMEM39A, GAB2, STAT5A, ADAM12, CUBN, MBNL1, NFKB2, ENOX1, IRAK4, ARHGEF2, CYLD, MCM5
dendritic | negative: ABCA10, ABCA8, ABCB1, ABCC12, ABCC8, ABCC9, ABI3, ABI3BP, AC007906.2, ACPP, ACVRL1, ADAM33, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRB2, ADGRG6, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADRA2A, ADTRP, AGMAT, AHRR, AIM2, ALDH1B1, ALPL, AMOT, ANGPT1, ANGPTL2, ANGPTL4, ANKRD6, ANXA3, ANXA8, APLNR, ARG2, ARL4D, ASPN, ATAD3A, ATL1, AUNIP, AURKB, AVPR1A, AXIN2, BCL6B, BDKRB2, BLM, BMP2, BMP6, BMPR1B, BNC2, BPIFB1, BRCA1, BRINP1, C1QTNF1, C1QTNF6, C3AR1, C4B, C5AR1, CABP4, CABYR, CACNA1B, CACNA2D1, CACNG4, CADM3, CALML3, CARD9, CAVIN2, CBS, CCDC136, CCDC141, CCL14, CCL2, CCN3, CCN4, CCNE1, CCR4, CD19, CD1C, CD1D, CD209, CD22, CD247, CD27, CD28, CD33, CD3G, CD40LG, CD72, CD79B, CDC25A, CDC6, CDCA2, CDH13, CDH5, CDH6, CDK18, CDKN2B, CDKN3, CEBPA, CEMIP2, CENPV, CEP112, CES1, CFC1, CHST8, CLDN5, CLEC12A, CLEC14A, CLEC5A, CLEC7A, CLGN, CMA1, CNN1, CNRIP1, CNTNAP1, COL10A1, COL4A3, COL4A4, COLQ, COMP, CPXM2, CR1, CRB2, CREB5, CSPG4, CTLA4, CTNNAL1, CTSG, CX3CL1, CXCL2, CXCR6, CYP27A1, DCLK1, DDR2, DDX28, DHODH, DIO2, DLEC1, DLK1, DLL4, DLX4, DMD, DNAH10, DNAH5, DNAH7, DNMT3B, DOC2A, DOK7, DPT, DPY19L2, DRC3, DSC3, DSG3, DTX1, DZIP1L, EBF2, ECRG4, EDN1, EDNRA, EFR3B, EGF, EGR2, ENPP3, EPHA4, EREG, ERG, ERP27, ERRFI1, ERVMER34-1, ESM1, ETV1, ETV5, F2R, F2RL3, F7, FAM107A, FAM117B, FAM161A, FAT2, FAT4, FBXO16, FCGBP, FCGR2A, FCGR2B, FCGR3A, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGF2, FGF7, FGFR4, FLT1, FLT3LG, FLT4, FMOD, FOLH1, FOLR2, FOXC1, FOXC2, FOXD2, FPR1, FREM1, FST, GABRD, GABRP, GHR, GJC1, GLI2, GLS2, GNAI1, GNAO1, GPER1, GPR34, GPR39, GPR4, GPRASP2, GPRC5B, GRIK4, GRM4, GRP, GUCY1A2, GYG2, GZMA, GZMK, HAS2, HBEGF, HCN2, HDC, HEY1, HEYL, HGF, HMCN1, HMGN5, HNMT, HOXA5, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS6ST2, HSD11B2, HSPA6, HSPB8, ICOS, IFITM1, IGF1, IL18, IL1RAP, IL1RL1, IL2RB, IL33, IL34, INHBA, IRX1, ITGA2B, ITGA7, ITGB1BP2, ITIH5, ITK, ITPKA, JAM2, KCNAB3, KCNC4, KCNF1, KCNH6, KCNJ8, KCNK15, KCNMB1, KCNN3, KCNN4, KDR, KIAA0408, KIT, KITLG, KLHL13, KLK10, KLK6, KLRD1, LAMB3, LATS2, LCK, LCN2, LDB3, LEF1, LGR4, LIF, LIFR, LILRB5, LIPE, LMO2, LOX, LPAR1, LPAR5, LRG1, LRRC15, LTBP1, LURAP1, MALL, MAP2, MAP3K7CL, MAPK11, MAPK12, MAPK8IP1, MC1R, MECOM, MERTK, MET, METTL18, MFAP5, MID2, MIPOL1, MLXIPL, MMP9, MMRN2, MNS1, MPDZ, MRC1, MRM3, MS4A1, MS4A2, MSR1, MSTN, MTNR1B, MUC16, MZF1, NACC2, NAIF1, NAT1, NBPF3, NECTIN3, NEDD4, NEIL3, NEK2, NEXN, NGFR, NLGN3, NLRP3, NMB, NOD2, NOS1AP, NOS3, NOSTRIN, NPNT, NPR1, NPY1R, NR5A2, NRN1, NT5E, NTRK2, OLFM1, OLFML3, OLR1, OPRD1, OSBP2, OSCP1, OSGIN1, OXTR, P2RX5, PALMD, PAMR1, PAX5, PBK, PCDH10, PCDH17, PCDH18, PCDH8, PDE2A, PDE3A, PDGFRA, PDGFRL, PDZK1, PGBD1, PGF, PGR, PHF21B, PI16, PILRA, PIMREG, PINK1, PKP2, PLA2G7, PLD1, PLEKHH2, PLIN1, PLK4, POTEI, POTEJ, POU2AF1, PPARG, PPFIA3, PPP1R1C, PPP2R3A, PRDM6, PRF1, PRICKLE1, PRKCQ, PRKD1, PRMT5, PROCR, PRODH, PROM1, PROS1, PROX1, PRRT2, PSRC1, PSTPIP2, PTAFR, PTGDR, PTGES, PTGIS, PTGS1, PTPRB, PTPRM, PYGL, QPCTL, RAD54L, RAPGEF3, RASGRP1, RBBP4, RCAN2, RECK, RELN, RFX2, RGS5, RNF180, RNF26, RNF39, ROBO4, ROR2, RPA3, RRAS, RTN1, RUNX1T1, SAC3D1, SAMD5, SATB2, SCGB1D2, SCIMP, SELP, SEPTIN5, SERPINB5, SESN2, SFRP4, SH2D1A, SH2D2A, SH3RF2, SHANK3, SIGLEC1, SIGLEC9, SIM2, SKA3, SLAMF8, SLC11A1, SLC16A9, SLC18A2, SLC1A3, SLC26A2, SLC2A3, SLC35G2, SLC6A1, SLC7A7, SLCO2A1, SLIT2, SLIT3, SOCS2, SOD3, SOSTDC1, SOX10, SOX17, SOX5, SPAG1, SPAG17, SPATA18, SPOCK1, SPON1, SSBP3, STAC3, STAP1, STEAP1, STMN3, STS, SUGCT, SULT1A1, SUV39H1, SVEP1, TAL1, TBC1D8B, TBX2, TBXA2R, TDO2, TEK, TEKT3, TEX30, TFAP2B, TFR2, TGFB2, TGFBR3, TGM2, TH, TIE1, TIGIT, TLE6, TLR5, TMEM119, TMEM255B, TMEM98, TNFRSF10C, TNFRSF13B, TNFRSF13C, TP63, TPMT, TPPP3, TPSG1, TRAF3IP2, TREM2, TRH, TRIB3, TRPC1, TRPC6, TSPAN18, TSPAN7, TTC23, TUBB2B, TWIST1, TWIST2, UBASH3A, UCN, UNC5A, VDR, VGLL3, VSIG4, VSNL1, VTCN1, WNT2, WNT4, ZBP1, ZFHX4, ZHX3, ZNF165, ZNF177, ZNF232, ZNF408, ZNF683
endo | positive: CDH5, PLVAP, ADGRL4, CALCRL, FLT1, KDR, PTPRB, CLEC14A, FLT4, DOCK9, ITGA5, TIE1, MMRN2, ROBO4, PTPRM, PRCP, PDGFB, S1PR1, SHANK3, SLCO2A1, TGFBR2, CLDN5, THBD, CCL14, NOSTRIN, EFNB2, ELK3, RAPGEF3, NEDD9, HOXD9, NPR1, APLNR, KLF4, TSPAN18, TEK, LMO2, PALMD, CDH13, MAPK11, PCDH17, NR5A2, ACVRL1, BCL6B, MECOM, LIFR, TGM2, PDE2A, ABL2, ICAM2, SOCS3, IL33, OLFM1, NRN1, JAM2, DLL4, SLC35G2, TSPAN7, GPRC5B, PLEKHA1, CEMIP2, SOCS2, EDN1, PROX1, HBEGF, SOX17, SELP, EPHA4, EPAS1, TMEM255B, TMEM173, CAVIN2, DUSP6, NOTCH1, IVNS1ABP, SPTBN1, IL3RA, BMPR2, GIMAP8, NDRG1, PPARG, MAPK12, ERG, PIEZO2, ANGPTL2, ADAMTS4, PTP4A3, MET, TGFBR3, VAMP5, FYN, FAT4, RGS5, ABCB1, CNRIP1, CPXM2, C1QTNF1, PRKCH, FAP, NOD1, TAL1, ABI3, IL1R1, GJC1, FBLN2, ZNF608, NOS3, HLX, LOX, F2RL3, FOLH1, HOXD10, KCNN3, BMP6, APP, TJP1, PLCG1, SNTB2, BCR, CTTNBP2NL, AKT3, DNAJB4, PNP, EGLN1, EGFL8, HOXB4, LATS2, TECPR1, APOL1, F2R, KCNC4, MAFF, GPR4, ESM1, RCAN2, CX3CL1, CCDC80, GABRD, BDKRB2, PROCR, FAM107A, NPNT, TBXA2R, BMP2, HEY1, RELN, CASP10, PICALM, MRTFB, NOL4L, CRYBG3, MSN, PML, GNAI1, ARHGEF7, PINK1, ABCD4, CUBN, MALL, CEP112, RUNX1T1, PGF, AKR1C3, ITGA7, FLI1, BCAR1, ECE1, TSPAN9, CDC37, CLIC4, DISC1, ATXN3, DLG4, ZHX3, RNF180, TPPP3, NECTIN3, TRPC6, FGF2, ITIH5, PROS1, MRC1, C7, ABCC9, TBX2, RECK, CARD8, APOL3, SH3BP5, IL6ST, ARRB1, PTK2, VAMP3, CAMSAP2, LRP6, UTRN, PLSCR1, MPDZ, LIPA, PVR, TTC23, HSPB8, SLC2A3, IFITM1, CDKL1, FOXO1, HHEX, NFATC1, LRCH1, NRROS, CDK17, MEF2A
endo | negative: ABCA10, ABCA8, ABCC12, ABCC8, AC007906.2, ACPP, ADAMDEC1, ADAMTS14, ADCYAP1, ADGRB1, ADGRE2, ADORA2B, ADORA3, ADRA1B, ADTRP, AGMAT, AHRR, ANXA8, APOBEC3D, ARG2, ARL4D, ATL1, AUNIP, BANK1, BCL2A1, BLK, BLM, BMPR1B, BPIFB1, BRCA2, BRINP1, C3AR1, C4B, C5AR1, CABYR, CACNG4, CADM3, CALB2, CALML3, CARD9, CBS, CCL22, CCN4, CCNE1, CCR1, CCR4, CCR5, CCR6, CD19, CD1C, CD1E, CD209, CD22, CD226, CD247, CD27, CD28, CD33, CD38, CD3G, CD40LG, CD5, CD7, CD72, CD79A, CD86, CD8A, CDC25A, CDC6, CDK18, CEBPA, CES1, CFC1, CHST8, CLEC10A, CLEC12A, CLEC4A, CLEC4C, CLEC5A, CLEC7A, CLGN, CMA1, CMKLR1, CNR2, COCH, COL10A1, COL4A3, COL4A4, CR1, CRB2, CSF3R, CST7, CTLA4, CTSG, CTSW, CUX2, CX3CR1, CXCR3, CXCR6, DERL3, DLEC1, DLK1, DLX4, DNAH10, DNAH5, DNAH7, DNM1, DOC2A, DOK7, DPT, DRC3, DSC3, DSG3, ECRG4, EGF, EGR2, ENPP3, EREG, ERVMER34-1, F7, FAM161A, FAT2, FBXO16, FCER1A, FCGR2A, FCGR2B, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGFR4, FLT3, FOLR2, FOXD2, FPR1, FREM1, FST, FUT7, GABRP, GLI2, GLS2, GPR174, GPR39, GRIK4, GRM4, GRP, GYG2, GZMA, GZMB, GZMK, HAS2, HAVCR2, HCK, HCN2, HDC, HGF, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, ICOS, IDO1, IL12RB1, IL18, IL18R1, IL1RL1, IL21R, IL2RB, IL6R, IRF4, IRF5, IRX1, ITGA2B, ITGB1BP2, ITK, ITPKA, KCNA3, KCNF1, KCNH6, KCNK10, KCNK15, KCNK17, KCNMA1, KCNN4, KIAA0408, KIT, KLHL13, KLK10, KLK6, KLRD1, KMO, LAMB3, LAMP3, LCN2, LCNL1, LDB3, LGR4, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LPAR1, LPAR5, LRG1, LTK, MAP1A, MAPK8IP1, MC1R, MELTF, MITF, MLXIPL, MNDA, MNS1, MS4A1, MS4A2, MSTN, MTNR1B, MUC16, NBPF3, NCCRP1, NCF1, NCF2, NEIL3, NEK2, NEK8, NIBAN3, NLGN3, NLRP3, NMB, NOD2, NOS1AP, NPY1R, NR4A3, NSUN5, NT5DC3, NTRK2, NXT2, OLR1, OPRD1, OSGIN1, P2RX1, P2RX5, P2RX7, P2RY13, P2RY14, PAX5, PCDH10, PCDH8, PDE3A, PDZK1, PGR, PHEX, PHF21B, PI16, PILRA, PIMREG, PKP2, PLA2G7, PLEKHH2, PLIN1, PLK4, PM20D2, POTEI, POTEJ, POU2AF1, POU2F2, PPFIA3, PPP1R1C, PRDM6, PRF1, PRKCQ, PRODH, PROM1, PSRC1, PSTPIP2, PTGDR, PTGES, PTGIS, PTGS1, RAD54L, RBBP4, RFX2, RIMS3, RNF39, ROR2, RPA3, RTN1, SCGB1D2, SCIMP, SCN9A, SDK2, SERPINB5, SESN2, SFRP4, SH2D1A, SH2D2A, SH3RF2, SHD, SIGLEC1, SIGLEC6, SIGLEC9, SIM2, SKA3, SLAMF6, SLAMF8, SLC11A1, SLC16A9, SLC18A2, SLC1A3, SLIT2, SMPD3, SOSTDC1, SOX10, SPAG17, SPATA18, SPIB, SPOCK1, SPON1, STAC3, STAP1, STEAP1, STX11, SUGCT, SULT1A1, SUV39H1, TBXAS1, TCL1A, TEKT3, TEX30, TFAP2B, TFR2, TH, TIGIT, TLE6, TLR1, TLR7, TLR9, TMEM200A, TMPRSS13, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFSF15, TNFSF8, TP63, TPSG1, TREM2, TREML2, TRH, TRPM2, TUBB2B, TWIST1, TWIST2, UBASH3A, VGLL3, VSIG4, VSNL1, VTCN1, WNT2, WNT4, WNT5B, ZBP1, ZFHX4, ZNF165, ZNF232, ZNF683
macro | positive: MS4A6A, CD163, CSF1R, C1QC, CD14, MAFB, AOAH, MSR1, TYMP, TLR2, SLCO2B1, LGMN, SIGLEC1, HSPA6, FCGR2A, CPVL, CLEC7A, GPR34, HAVCR2, AIF1, CSF3R, DMXL2, IFNGR1, CREG1, MRC1, NCOA4, LIPA, F13A1, FCGR3A, NCF2, VSIG4, SLAMF8, SLC40A1, CLEC10A, ATG7, FOLR2, MPP1, AKR1B1, AP1B1, IL18, SIRPA, FKBP15, P2RY13, EPB41L3, LILRB2, SLC7A7, MERTK, PLA2G7, HMOX1, FGR, NLRP3, LY96, FUCA1, SOAT1, LILRB5, MNDA, TRPM2, TREM2, TLR1, C5AR1, CD209, PILRA, SLC1A3, P2RX7, OLR1, CD86, FCGR2B, MKNK1, CCR1, CD1D, SNX10, CSF2RA, C3AR1, LAIR1, CTSC, DISC1, KCNMA1, MAN2B1, LGALS9, HEXA, HCK, SNX30, PLEKHO1, PLAUR, ADORA3, NR1H3, PTAFR, MEF2A, JAK2, SLC43A2, CASP1, MYO5A, ABI3, GIMAP8, ADGRE2, PTGS1, FPR1, SLC11A1, TNFSF13B, TLR7, SIGLEC9, CEBPA, HNMT, CARD9, APAF1, CD33, RTN1, FCGBP, CLEC4A, NRROS, CLEC12A, CLEC5A, HPSE, CD1C, CR1, ACP5, METRNL, AP2A2, PTPRJ, TPCN1, CMKLR1, TBXAS1, SLC2A9, OLFML3, IFI44L, PSTPIP2, P2RY6, IL17RA, LILRB4, MMP9, FCER1A, ADAMDEC1, CD1E, GBP5, BCL2A1, SCIMP, CD72, LPAR5, PFKFB3, ALDH2, ZSWIM6, ARMH3, PLSCR1, CD302, PRCP, KCNMB1, TNFSF12, PLEKHM2, WDR81, TLR5, DRAM1, SATB1, HPS5, ETV5, IL6R, CREB5, GAB2, HGF, LACC1, CX3CR1, LMO2, STX11, NFATC2, ARRDC3, CD38, IRF5, ARRB1, PICALM, TGFBR1, CTSA, SLC11A2, IFNGR2, CORO1C, PDE4DIP, CEP170, CLCN5, MITF, ACPP, CCR5, CD28, TNFRSF14, SNX29, CD58, RAP1A, POU2F2, NAAA, PER1, HEXB, IVNS1ABP, PNPLA6, P4HA1, NAIP, JDP2, CAMK1, CBL, FLT3, IGF1, STAT5A, PTPN6, FLI1, PLEKHA2, SLC23A2, SERPINB9, CARD8, IFNAR2, CD226, MX1, ADAM8, IL10RB
macro | negative: ABCA10, ABCA8, ABCB1, ABCC12, ABCC8, ABCC9, ABI3BP, ABTB2, AC007906.2, ACVRL1, ADAM12, ADAM33, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRB1, ADGRB2, ADGRL4, ADORA2B, ADRA1B, ADRA2A, ADTRP, AGMAT, AHRR, AKR1C3, ALPL, ANGPT1, ANGPTL2, ANGPTL4, ANXA3, ANXA8, APCDD1, APLNR, APOBEC3D, ARG2, ARL4D, ASPN, ATL1, AUNIP, AVPR1A, AXIN2, BCL6B, BDKRB2, BLK, BLM, BMP2, BMP6, BMPR1B, BPIFB1, BRINP1, C1QTNF1, C4B, C7, CABYR, CACNA2D1, CACNG4, CADM3, CALB2, CALCRL, CALML3, CAVIN2, CBS, CCDC136, CCL14, CCL19, CCL22, CCL28, CCN3, CCN4, CCNE1, CD19, CD27, CD79A, CD79B, CD8A, CDC25A, CDH13, CDH5, CDH6, CDKL1, CDKN2B, CENPV, CEP112, CFC1, CHST8, CLDN5, CLEC14A, CLEC4C, CMA1, CNN1, CNR2, CNTNAP1, COCH, COL10A1, COL4A3, COL4A4, COLQ, COMP, CPXM2, CRB2, CSPG4, CTSG, CTSW, CUBN, CUX2, CX3CL1, CXCR6, DCLK1, DDR2, DERL3, DHODH, DIO2, DLEC1, DLK1, DLL4, DLX4, DNAH10, DNAH5, DNAH7, DNMT3B, DOK7, DPT, DPY19L2, DRC3, DSC3, DSG3, DTX1, EBF2, ECRG4, EDN1, EDNRA, EFR3B, EGF, ENOX1, ENPP3, EPHA4, EPHB1, EREG, ERG, ERP27, ERVMER34-1, ESM1, ETV1, F2R, F2RL3, F7, FAM107A, FAM161A, FAP, FAT2, FAT4, FBXO16, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGF2, FGF7, FGFR4, FLT1, FLT4, FMOD, FOLH1, FOXC1, FOXC2, FOXD2, FREM1, FST, GABRD, GABRP, GHR, GJC1, GLI2, GLS2, GNAO1, GPR39, GPR4, GPRASP2, GPRC5B, GRIK4, GRM4, GRP, GUCY1A2, GXYLT2, GYG2, GZMB, GZMK, HAS2, HCN2, HCN3, HDC, HEY1, HEYL, HMCN1, HMGN5, HOXA5, HOXB4, HOXD10, HOXD9, HPGD, HPSE2, HS3ST3A1, HSD11B2, HSPB8, ICOS, IFITM1, IL18R1, IL1RL1, IL33, IL34, IL3RA, INHBA, IRF4, IRX1, ITGA2B, ITGA7, ITGB1BP2, ITIH5, ITPKA, JAM2, KCNAB3, KCNC4, KCNF1, KCNH6, KCNJ8, KCNK10, KCNK15, KCNK17, KCNN3, KDR, KIAA0408, KIT, KITLG, KLHL13, KLK10, KLK6, KLRD1, LAMB3, LCN2, LCNL1, LDB3, LEF1, LGR4, LIF, LILRA4, LIPE, LOX, LRG1, LRRC15, LTBP1, LTK, LZTS1, MAGI3, MALL, MAP1A, MAP2, MAP3K7CL, MAPK11, MAPK8IP1, MC1R, MECOM, MED12L, MET, MFAP5, MLXIPL, MMP11, MMRN2, MNS1, MRM3, MS4A1, MS4A2, MSTN, MTNR1B, MUC16, NBPF3, NCCRP1, NECTIN3, NEIL3, NEK8, NEXN, NGFR, NIBAN3, NLGN3, NMB, NOS1AP, NOS3, NOSTRIN, NPNT, NPR1, NPY1R, NR5A2, NRN1, NT5E, NTRK2, OPRD1, OSBP2, OSCP1, OSGIN1, OXTR, P2RX5, PALMD, PAMR1, PAX5, PCDH10, PCDH17, PCDH18, PCDH8, PDE2A, PDE3A, PDGFRA, PDGFRL, PDZK1, PGAP1, PGBD1, PGF, PGR, PHEX, PHF21B, PHLPP1, PI16, PIEZO2, PIMREG, PKP2, PLEKHH2, PLIN1, PLK4, POTEI, POTEJ, POU2AF1, PPFIA3, PPP1R1C, PPP2R3A, PRDM6, PRF1, PRICKLE1, PRKCQ, PRKD1, PRODH, PROM1, PROS1, PROX1, PRRT2, PTCH1, PTGDR, PTGES, PTGIR, PTGIS, PTP4A3, PTPRB, RASD1, RBBP4, RCAN2, RECK, RELN, RFX2, RGS5, RIMS3, RNF180, RNF39, ROBO4, ROR2, RPA3, RUNX1T1, S1PR1, SAMD5, SATB2, SCGB1D2, SDK2, SELP, SEMA7A, SEPTIN5, SERPINB5, SESN2, SFRP4, SH2D2A, SH3RF2, SHANK3, SHD, SIGLEC6, SIM2, SKA3, SLC16A9, SLC18A2, SLC35G2, SLC6A1, SLCO2A1, SLIT2, SLIT3, SMPD3, SOCS2, SOD3, SOSTDC1, SOX10, SOX17, SOX5, SPAG1, SPAG17, SPATA18, SPOCK1, SPON1, SRPX, STAP1, STEAP1, STMN3, STRIP2, SUGCT, SUV39H1, SVEP1, SYNCRIP, TAL1, TBC1D8B, TBX2, TBXA2R, TCL1A, TDO2, TEK, TEKT3, TFAP2B, TFR2, TGFB2, TGFBR3, TH, THBS2, TIE1, TIGIT, TLE6, TLR9, TMEM200A, TMEM98, TMPRSS13, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TP63, TPPP3, TPSG1, TRH, TRIB3, TRPC1, TSPAN18, TSPAN7, TUBB2B, TWIST1, TWIST2, UBASH3A, UCN, VGLL3, VSNL1, VTCN1, WFDC1, WNT2, WNT4, ZFAT, ZFHX4, ZNF165, ZNF232, ZNF408, ZNF608, ZNF683
mast | positive: KIT, HDC, IL1RL1, MS4A2, SLC18A2, FOXP1, CTSG, HPGD, TPSG1, FER, HPGDS, ADCYAP1, KLRG1, STXBP5, IL18R1, SIGLEC6, LIF, P2RX1, PTGS1, CMA1, ACSL4, ADGRE2, ENPP3, ABCC1, MCTP2, AGAP1, LAT, CAVIN2, TMEM255B, PLAUR, C3AR1, CD274, CD22, CD33, CD82, PPM1H, AP1S3, CALB2, NEK6, ABCC4
mast | negative: AAR2, ABCA10, ABCB1, ABCB6, ABCC12, ABCC9, ABCD4, ABI3, ABI3BP, ABL2, ABTB2, AC007906.2, ACACA, ACKR3, ACLY, ACP5, ACVRL1, ADA, ADAM33, ADAMTS4, ADGRB2, ADGRG6, ADGRL4, ADIPOR2, ADORA2B, ADRA1B, ADTRP, AHRR, AIF1, AIM2, AKT2, ALDH1B1, ALDH2, ALDH7A1, ALPK1, AMACR, AMOT, ANGPTL2, ANGPTL4, ANKRD13A, ANKS6, ANXA3, AOAH, APAF1, APCDD1, APEX1, APEX2, APLNR, APOL1, APOL3, APTX, ATAD3A, ATL1, ATP6V1H, ATPAF1, AURKA, AURKB, AVPR1A, AXIN2, BAG3, BANK1, BARD1, BCAR3, BCCIP, BCL11B, BCL2L12, BCL6B, BLK, BLM, BLOC1S2, BMPR1B, BMPR2, BPIFB1, BRCA1, BRCA2, BSDC1, BTRC, C1QBP, C1QC, C1QTNF1, C1QTNF6, C5AR1, CABP4, CACNA1B, CACNA1D, CACNA2D1, CACNG4, CALCRL, CALML3, CAMSAP2, CARM1, CASP1, CBL, CBS, CCDC124, CCDC137, CCDC170, CCDC50, CCDC80, CCL14, CCL28, CCN4, CCNE1, CCR1, CCT2, CD14, CD163, CD19, CD1D, CD2, CD209, CD247, CD27, CD28, CD320, CD3D, CD3E, CD3G, CD5, CD6, CD79A, CD79B, CD86, CD8A, CD96, CDAN1, CDC25A, CDC6, CDCA2, CDH13, CDH23, CDH5, CDH6, CDK18, CDK4, CDK7, CDKN1A, CDKN2B, CDKN3, CEBPA, CEMIP2, CENPF, CENPV, CEP170, CEP85, CES1, CIP2A, CISD2, CKAP2, CLCN5, CLDN5, CLEC10A, CLEC14A, CLEC4C, CLEC7A, CLGN, CLIC4, CLSTN3, CMKLR1, CMSS1, CNIH4, CNN1, CNNM3, CNP, COLQ, COMP, COPS5, CORO1C, CPVL, CPXM2, CR1, CRYBG1, CRYBG3, CSE1L, CSF1R, CSF2RA, CSF3R, CSNK2A1, CSPG4, CTDSPL, CTNNA1, CTNNAL1, CTSC, CTSK, CX3CL1, CXCR3, CYCS, CYHR1, CYP1B1, CYP24A1, CYP27A1, DAG1, DBN1, DCLK1, DDIT3, DDX21, DDX28, DERL3, DGAT1, DHODH, DHX30, DHX34, DHX57, DIO2, DLEC1, DLK1, DLL4, DLX4, DMD, DMXL1, DNAH5, DNAH7, DNAJA1, DNAJB4, DNASE1L3, DNM1, DNMT3B, DOCK9, DPAGT1, DPT, DSC3, DSG3, DUSP5, DVL1, DVL2, DZIP1L, E2F3, EBF2, ECRG4, ECSIT, EDN1, EDNRA, EEF1AKNMT, EFNB2, EFR3B, EGF, EGFL8, EGLN3, EHMT2, EIF2AK3, EIF2S1, EIF4ENIF1, EIPR1, ELK1, ELK3, ELOC, EPB41L3, EPHA4, EPHB1, EPHX1, ERCC2, EREG, ERLIN2, ERRFI1, ERVMER34-1, ETV1, F13A1, F2R, F7, FADD, FAF2, FAM117B, FAM53C, FAM98A, FANCC, FANCI, FAP, FAS, FAT2, FAT4, FBLN2, FBXO16, FBXO25, FBXW11, FCGR2A, FCGR2B, FCGR3A, FCMR, FCRL1, FCRL2, FCRL5, FDPS, FECH, FGF1, FGF7, FGFR4, FGR, FH, FHL2, FIBCD1, FIBP, FLT1, FLT3, FLT4, FMOD, FN3KRP, FOLR2, FOXC1, FOXO1, FRMD6, FST, FUCA1, FUT7, FXR2, FZD1, GABRP, GADD45A, GADD45G, GALNT4, GART, GBP5, GCLM, GCNT2, GHR, GIMAP8, GJC1, GLA, GLS2, GMDS, GMFB, GMPS, GNA11, GNAO1, GPER1, GPN1, GPR137, GPR39, GPRASP2, GPRC5B, GRAMD2B, GRIK4, GRM4, GSTZ1, GUCY1A2, GXYLT2, GYG2, GZMA, GZMB, HAX1, HBEGF, HCK, HCN2, HCN3, HDAC2, HDAC3, HELLS, HEYL, HGH1, HLX, HMCN1, HMGA1, HMGN5, HMOX1, HNMT, HOXD9, HPS5, HS6ST2, HSD11B2, HSPA4, HSPA6, HSPD1, HTATIP2, ICAM2, ICAM3, IDO1, IFI44L, IGF1, IKBKE, IKZF4, IL17RA, IL1R1, IL2RB, IL33, IL34, IL3RA, IL6R, IL7R, IMPDH1, INHBA, IPO5, IRF1, IRF2BP1, IRF2BPL, IRF4, IRX1, ITGA5, ITGA7, ITGB1BP2, ITGB5, ITK, ITPR3, JAM2, JDP2, KCNC4, KCNH6, KCNJ8, KCNK15, KCNK17, KCNMB1, KCNN4, KDR, KEAP1, KITLG, KLF12, KLF16, KLF4, KLK6, KLRK1, KMO, LAMB3, LAMP3, LANCL2, LATS2, LCAT, LCK, LCNL1, LGALS9, LGMN, LGR4, LIFR, LIG4, LILRA4, LILRB5, LIPA, LIPE, LMO2, LOX, LOXL1, LPAR1, LRG1, LRP6, LRRC59, LSM12, LTBP1, LTBP2, LTK, LY96, MAEA, MAFB, MAFF, MAGI3, MANEA, MAP1A, MAP2K4, MAP3K14, MAP3K20, MAP4K3, MAPK11, MC1R, MCM3, MCM6, MDFIC, MECOM, MED12L, MEIS1, MEPCE, MERTK, MET, METTL18, MFAP5, MGMT, MID2, MINDY2, MIPOL1, MLF1, MLXIPL, MMP11, MMRN2, MNDA, MNS1, MPDU1, MPDZ, MRC1, MRFAP1L1, MRM3, MRTFB, MS4A1, MS4A6A, MSH2, MSH3, MSR1, MSTN, MTCH2, MTERF2, MTNR1B, MUC16, MX1, MYBBP1A, MYBL2, MYLIP, MYO1C, MYOF, N4BP2, NAIF1, NAIP, NAMPT, NAT1, NAT10, NAV2, NBL1, NBPF3, NCF2, NDRG1, NEDD4, NEK2, NEK4, NEK8, NGFR, NIBAN3, NIFK, NLRP3, NMB, NOA1, NOD1, NOP2, NOS1AP, NOSTRIN, NOTCH3, NPC1, NPR1, NPY1R, NQO2, NR1H3, NR5A2, NRN1, NSUN5, NT5DC3, NT5E, NTPCR, NTRK2, NUCB2, NUDCD2, NUDT1, NXT2, OLFM1, OLFML3, OLR1, OPRD1, ORAI2, OSBP2, OSCP1, OSGIN1, OXSM, OXTR, P2RX5, P2RX7, P2RY13, PALMD, PAMR1, PARP1, PAX5, PBDC1, PBK, PCDH10, PCDH17, PCDH18, PDE2A, PDE4DIP, PDGFB, PDGFRA, PDGFRL, PDK1, PDLIM2, PDZK1, PELO, PGAP1, PGBD1, PGD, PGF, PGM1, PHB2, PHEX, PHF21B, PHLPP1, PIEZO2, PILRA, PIMREG, PINK1, PLA2G7, PLCG1, PLD1, PLEKHA1, PLEKHH2, PLIN1, PLK4, PLVAP, PM20D2, PNP, PNPT1, POLDIP2, POLE, POMGNT1, POMT1, POR, POTEI, POU2AF1, POU2F2, PPARG, PPID, PPP1R1C, PPP2R3A, PRCP, PRDM1, PRDM10, PRDM6, PRICKLE1, PRKCH, PRKCI, PRKCZ, PRKD1, PRMT5, PRODH, PROM1, PROX1, PRPS2, PRRT2, PSEN2, PSENEN, PSMD4, PSRC1, PTGES, PTGFRN, PTGIS, PTK7, PTP4A3, PTPN11, PTPRB, PTPRJ, PTPRM, PVR, PYCARD, R3HDM1, RAB11FIP1, RAB27A, RAC1, RAD18, RAD54L, RAPGEF3, RASD1, RASGRP1, RAVER1, RBM10, RBM38, RCAN2, REEP4, RELB, RFC4, RGS5, RIPK2, RIPK3, RITA1, RMND1, RNF121, RNF26, ROBO4, ROR2, RPA3, RRAS, RRM1, RRP15, RUBCN, RUNX1T1, RUNX2, RUVBL1, RWDD4, SAC3D1, SAMD5, SCAF4, SCGB1D2, SCIMP, SCN9A, SDHAF3, SELP, SEPTIN5, SERPINB5, SESN2, SF3B4, SFRP4, SFXN1, SH3BP5, SH3RF2, SHANK3, SHARPIN, SHD, SHLD1, SHMT1, SHMT2, SIAE, SIGLEC1, SIGMAR1, SIM2, SIRPA, SLAMF1, SLAMF8, SLC16A1, SLC1A3, SLC20A2, SLC25A1, SLC29A1, SLC2A9, SLC35G2, SLC37A4, SLC39A9, SLC43A2, SLC6A1, SLC7A5, SLC7A7, SLCO2A1, SLIT2, SLIT3, SMPD3, SNX10, SNX15, SNX27, SNX30, SNX8, SOAT1, SOCS2, SOCS3, SOD3, SOSTDC1, SOX10, SOX17, SOX2, SOX5, SPAG17, SPAG5, SPATA18, SPIB, SPOCK1, SPON1, SRA1, SRC, SRPX, SSBP3, SSX2IP, STAC3, STAT4, STEAP1, STK16, STMN1, STMN3, STS, SULT1A1, SUV39H1, SVEP1, SVIL, SYNCRIP, SYVN1, TBRG4, TBX2, TBX3, TCF7, TCF7L2, TCFL5, TCL1A, TCP1, TECPR1, TEK, TEX30, TFAM, TFAP2B, TFPI2, TGFBR3, TGM2, THBD, THBS2, THEMIS, THYN1, TICAM1, TIPARP, TIRAP, TJP1, TLR1, TLR2, TLR5, TLR7, TLR9, TMEM119, TMEM120B, TMEM192, TMEM97, TMEM98, TMPRSS13, TNFRSF13B, TNFRSF13C, TNFRSF6B, TNFSF13B, TOLLIP, TOR3A, TP63, TPMT, TPPP3, TRAC, TRAF1, TRAFD1, TRBC1, TREM2, TRIB3, TRIM28, TRPC6, TRPM2, TRPV1, TRUB1, TSPAN18, TSPAN9, TSPYL5, TTC12, TTC23, TUBG1, TWIST1, TWIST2, UBAP2, UBE2T, UCN, UCP2, UFC1, UNC5A, URB1, URGCP, USP5, VAMP5, VDR, VSIG4, VSNL1, VTCN1, WASL, WDHD1, WDR12, WDR55, WFS1, YAP1, YIPF1, YIPF5, YJEFN3, YTHDF1, ZAP70, ZBTB10, ZBTB33, ZC3H12A, ZFAT, ZFYVE26, ZGRF1, ZHX3, ZNF232, ZNF281, ZNF408, ZNF608, ZNF687, ZNF827, ZSWIM6
myoepi | positive: DST, CSRP1, ERRFI1, IRX1, GRAMD2B, TP63, LAMB3, DSC3, ANXA3, FAT2, NAV2, CALML3, SAMD5, SOX10, CX3CL1, PROM1, FST, ALDH7A1, GABRP, GCLM, CCL28, NGFR, SERPINB5, DSG3, VTCN1, LGR4, ECRG4, IL34, SH3RF2, OXTR, KLK6, CES1, SOSTDC1, ADTRP, ADORA2B, TFAP2B, FGF1, VSNL1, AC007906.2, LTBP2, FHL2, FRMD6, CNN1, NTRK2, DMD, KCNMB1, TLR2, CSPG4, MET, SLC2A9, CDH13, PPP2R3A, NEDD9, TMEM98, FOXC1, VDR, COMP, ZNF608, PDGFRA, KIT, KLHL13, ATL1, IL1RAP, TUBB2B, CADM3, LCN2, GRP, BRINP1, CXCL2, CRB2, RNF39, KLK10, ANXA8, KCNN4, RUNX1, YAP1, PIK3R1, SVIL, NDRG2, KCNMA1, FZD1, KCNC4, MYBBP1A, PSTPIP2, PAMR1, CDK18, NT5DC3, GPRC5B, FMOD, ETV1, MELTF, PALMD, HOXA5, TGFB2, PGR, EDN1, MUC16, WNT4, PKP2, ADGRB1, TCF7, TTC12, APP, MYOF, DUSP6, SLMAP, PTK7, PTGFRN, PLD1, MAFF, BCAR3, CYP27A1, CDH23, GCNT2, ANGPTL4, NAT1, CCDC136, RFX2, SPATA18, DIO2, CCN3, NOS1AP, NEXN, KCNK17, PTGES, PRICKLE1, TREM2, MMRN2, MAP2, PM20D2, SDK2, C1RL, TMEM43, PAPSS1, GADD45A, IL4R, SIRPA, TTC23, STMN3, APCDD1, RNF180, PROS1, ADAMTS5, SIRT3, FGF2, ETV5, INHBA, SVEP1, HMCN1, MAPK8IP1, TRPC1, CASP1, ANXA2, LAMP2, SRC, POMGNT1, ALDH3A2, SEPTIN5, DZIP1L, MPDZ, TIPARP, CTNNAL1, ALPK1, LCAT, SSBP3, ATF3, SOCS3, CEMIP2, TRAF3IP2, AMOT, DNAH7, SLC16A1, HSPB8, TLR5, SLC26A2, ZNF177, LURAP1, PYGL, AXIN2, CREB5, CCL2, ANKRD6, LRG1, PTCH1, CRYBG1, GABBR1, HPS5, PGM1
myoepi | negative: ABCA8, ABCC12, ABCC8, ABCC9, ABI3BP, ACVRL1, ADA, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADCYAP1, ADGRL4, ADORA3, ADRA1B, AGMAT, AIM2, APLNR, ASPN, AVPR1A, BANK1, BCL2A1, BCL6B, BLK, BMP2, BMP6, BNC2, C4B, C7, CALCRL, CCDC141, CCL14, CCL19, CCN4, CCR1, CCR4, CCR6, CD19, CD1C, CD1D, CD1E, CD209, CD22, CD226, CD27, CD274, CD28, CD33, CD38, CD3G, CD40LG, CD5, CD7, CD72, CD79A, CD79B, CD86, CDH6, CDKL1, CFC1, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4C, CLEC5A, CMA1, CMKLR1, CNR2, CNRIP1, CNTNAP1, COCH, COL10A1, COL4A3, COL4A4, CR1, CST7, CTLA4, CTSG, CUX2, CX3CR1, CXCR3, CXCR6, CYSLTR1, DERL3, DLEC1, DLK1, DLL4, DLX4, DNAH10, DNASE1L3, DPT, DTX1, EBF2, ENOX1, ENPP3, EPHB1, ESM1, F2RL3, FAM107A, FAT4, FCER1A, FCGR2B, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF7, FLI1, FLT1, FLT4, FOXD2, FPR1, FREM1, FUT7, GABRD, GIMAP8, GJC1, GLI2, GPR174, GPR4, GRIK4, GRM4, GUCY1A2, GZMA, GZMB, GZMK, HAS2, HCK, HDC, HEYL, HGF, HLX, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, ICAM2, ICOS, IDO1, IGF1, IL12RB1, IL18R1, IL1RL1, IL21R, IL3RA, IRF4, IRF5, ITPKA, KCNA3, KCNAB3, KCNF1, KCNJ8, KCNK10, KCNN3, KLRD1, KLRG1, LAMP3, LCNL1, LDB3, LILRA4, LILRB2, LILRB4, LILRB5, LPL, LRRC15, LTK, LY96, MAP1A, MAP3K7CL, MED12L, MFAP5, MNDA, MRC1, MS4A1, MS4A2, NCF1, NEIL3, NIBAN3, NLGN3, NLRP3, NOD2, NOS3, NPR1, NR4A3, NR5A2, NRN1, NRROS, OLR1, OPRD1, P2RX1, P2RX5, P2RX7, P2RY13, P2RY14, PAX5, PCDH17, PCDH8, PDE2A, PDE3A, PHEX, PI16, PIEZO2, PILRA, PIMREG, POTEJ, POU2AF1, PPP1R1C, PRDM6, PRF1, PTGDR, PTGIR, PTGIS, PTPRB, RCAN2, RELN, RGS5, ROBO4, RUNX1T1, S1PR1, SCIMP, SCN9A, SELP, SEMA7A, SFRP4, SH2D1A, SH2D2A, SHANK3, SHD, SIGLEC6, SIGLEC9, SLAMF6, SLC11A1, SLC18A2, SLC2A3, SLC6A1, SLIT2, SMPD3, SOD3, SOX17, SPIB, SPOCK1, SPON1, STAP1, STRIP2, STX11, TAL1, TBXA2R, TCL1A, TDO2, TEK, TFR2, TH, TIE1, TIGIT, TLR9, TMEM119, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFSF8, TPSG1, TREML2, TRH, TSPAN18, TSPAN7, TWIST1, TWIST2, UBASH3A, VSIG4, WFDC1, WNT2, ZBP1, ZFHX4, ZNF683
perivas | positive: NOTCH3, RGS5, GJC1, ITGA7, TBX2, CSPG4, TRPC6, ABCC9, CDH6, TPPP3, DPY19L2, HEYL, EDNRA, C1QTNF1, MAP3K20, SEPTIN5, KCNJ8, GUCY1A2, AVPR1A, EBF2, PGF, SLC6A1, SOX5, ADRA1B, EPAS1, SPECC1, ADAMTS4, SLIT3, PTP4A3, PCDH18, SOD3, PPARG, RCAN2, PDE3A, CNTNAP1, MAP2, ANGPT1, LDB3, CCND2, TDO2, NPNT, ADAMTS14, FLT1, MAP3K7CL, ARHGEF7, PLVAP, PAMR1, NOTCH1, DDR2, WFDC1, PELO, NT5DC2, GNAI1, PIEZO2, DMD, ADRA2A, CALCRL, THBS2, FOXC2, NDRG2, LPL, PTGIR, CDH5, KDR, LZTS1, KLF12, FGF7
perivas | negative: ABCA10, ABCC12, ABCC8, ABI3BP, ABTB2, AC007906.2, ACPP, ADAMDEC1, ADCYAP1, ADGRB1, ADGRE2, ADGRG6, ADORA2B, ADORA3, ADTRP, AGMAT, AIF1, AIM2, ANXA8, ARL4D, ATL1, AUNIP, BANK1, BCL11B, BCL2A1, BCL6B, BDKRB2, BLK, BLM, BMP6, BMPR1B, BNC2, BPIFB1, BRCA2, BRINP1, C3AR1, C4B, CACNA1B, CACNG4, CADM3, CALML3, CARD9, CASP10, CBS, CCL22, CCL28, CCNE1, CCR1, CCR4, CCR6, CD19, CD1C, CD1D, CD1E, CD209, CD22, CD27, CD274, CD33, CD38, CD7, CD72, CD79A, CD79B, CD86, CD8A, CDC25A, CDH13, CDH23, CDKN2B, CDKN3, CEBPA, CES1, CFC1, CHST8, CLEC10A, CLEC12A, CLEC4A, CLEC4C, CLEC5A, CLEC7A, CLGN, CMA1, CMKLR1, CNR2, COL10A1, COL4A3, COL4A4, COLQ, COMP, CPVL, CPXM2, CR1, CRB2, CREB5, CSF2RA, CSF3R, CST7, CTSG, CTSW, CUBN, CUX2, CX3CL1, CXCL2, CXCR3, CXCR6, CYP27A1, CYSLTR1, DDX28, DERL3, DLEC1, DLK1, DLL4, DLX4, DNAH10, DNAH5, DNAH7, DNASE1L3, DOC2A, DOK7, DPT, DRAM1, DRC3, DSC3, DSG3, ECRG4, EDN1, EFR3B, EGF, EGLN3, EGR2, ENPP3, EPB41L3, EPHA4, EPHB1, EREG, ERG, ERP27, ERVMER34-1, ETV1, F2RL3, F7, FAM117B, FAT2, FAT4, FBXO16, FCER1A, FCGR2A, FCGR2B, FCGR3A, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGFR4, FGR, FLT3, FLT4, FOLH1, FOLR2, FOXC1, FOXD2, FPR1, FREM1, FST, FUT7, GABRP, GADD45G, GBP5, GLI2, GLS2, GPER1, GPR174, GPR34, GPR39, GRIK4, GRM4, GRP, GYG2, GZMA, GZMB, GZMK, HAVCR2, HBEGF, HCK, HCN2, HDC, HHEX, HMCN1, HMGA1, HMGN5, HMOX1, HOXA5, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, HSPA6, ICAM3, IDO1, IGF1, IL12RB1, IL18, IL18R1, IL1RAP, IL1RL1, IL33, IL3RA, IL6R, IRF4, IRF5, IRX1, ITGB1BP2, ITK, ITPKA, JAM2, KCNAB3, KCNF1, KCNH6, KCNK10, KCNK15, KCNMA1, KCNN3, KCNN4, KIAA0408, KIT, KLHL13, KLK10, KLK6, KLRD1, KLRG1, KMO, LAMP3, LCK, LCN2, LCNL1, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LOX, LPAR1, LPAR5, LRG1, LRRC15, LTK, MAP1A, MAPK11, MAPK8IP1, MC1R, MED12L, MELTF, MET, MFAP5, MLXIPL, MNDA, MNS1, MRC1, MRM3, MS4A1, MS4A2, MSR1, MSTN, MTNR1B, MUC16, NBPF3, NCF1, NCF2, NEIL3, NEK2, NEK8, NFATC1, NIBAN3, NLGN3, NLRP3, NMB, NOD2, NOS1AP, NOS3, NPR1, NR4A3, NR5A2, NRROS, NSUN5, NXT2, OLR1, OPRD1, OSBP2, OSCP1, OSGIN1, OXSM, OXTR, P2RX1, P2RX5, P2RX7, P2RY13, P2RY6, PAX5, PCDH10, PCDH17, PCDH8, PDE2A, PDGFRA, PDGFRL, PDZK1, PGBD1, PHEX, PHF21B, PHLPP1, PI16, PILRA, PIMREG, PKP2, PLA2G7, PLAUR, PLIN1, PLK4, POTEI, POTEJ, POU2AF1, POU2F2, PPFIA3, PPP1R1C, PRDM6, PRF1, PRICKLE1, PRKCQ, PRODH, PROM1, PROX1, PSTPIP2, PTAFR, PTGES, PTGIS, PTGS1, QPCTL, RAD54L, RBBP4, RECK, RELN, RIMS3, RIPK3, RNF39, ROBO4, ROR2, RTN1, SAC3D1, SAMD5, SCGB1D2, SCIMP, SCN9A, SDHAF3, SDK2, SELP, SEMA7A, SERPINB5, SFRP4, SH2D1A, SH2D2A, SH3RF2, SHANK3, SHD, SIGLEC1, SIGLEC6, SIGLEC9, SIM2, SIRPA, SLAMF6, SLAMF8, SLC11A1, SLC16A9, SLC18A2, SLC1A3, SLC35G2, SLC7A7, SLCO2A1, SMPD3, SNX10, SOSTDC1, SOX10, SOX17, SPAG1, SPAG17, SPATA18, SPIB, SPOCK1, SPON1, STAC3, STAP1, STAT5A, STEAP1, STRIP2, SYNCRIP, TAL1, TBXAS1, TCL1A, TEK, TEKT3, TEX30, TFAP2B, TFPI2, TFR2, TGM2, TICAM1, TIE1, TIGIT, TLR1, TLR2, TLR7, TLR9, TMEM255B, TMPRSS13, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFSF13B, TNFSF8, TP63, TPMT, TPSG1, TRAF3IP2, TREM2, TRH, TRPM2, TSPAN18, TSPAN7, TUBB2B, TWIST2, UBASH3A, UCN, VDR, VSIG4, VSNL1, VTCN1, WNT2, WNT4, YTHDF1, ZBP1, ZC3H12A, ZFHX4, ZNF165, ZNF177, ZNF683
stromal | positive: THBS2, CCDC80, CTSK, FBLN2, ADAM12, FAP, PDGFRA, LOXL1, NBL1, ITGB5, DNM1, ADAM33, HMCN1, OLFML3, PLEKHH2, SVEP1, TMEM119, SRPX, ABCA10, ABI3BP, CLSTN3, F2R, FREM1, PTGFRN, MEIS1, FGF7, IGF1, SPON1, PDGFRL, LPAR1, GXYLT2, SLIT2, RUNX1T1, CYP1B1, MFAP5, INHBA, MMP11, PRICKLE1, FMOD, DCLK1, CCN4, SPOCK1, TWIST2, STEAP1, PTGIS, DPT, CDKN2B, SFRP4, LTBP2, FRMD6, PCDH18, ANGPTL2, MAFB, PTK7, IL1R1, APOL1, BNC2, LOX, SLIT3, COMP, ROR2, CPXM2, ANGPTL4, DIO2, TWIST1, ZFHX4, RECK, WNT2, CACNA2D1, COL10A1, VDR, HPSE2, PI16, LRRC15, GLI2, EGR2, SVIL, TCF7L2, PDE4DIP, FLCN, DRAM1, DDR2, SOD3, ASPN, TRPC1, RELN, CCL2, VGLL3, HAS2, SUGCT, ABCA8, DTX1, COL4A4, STAT2, SPART, FAS, STMN3, PTPRM, RUNX2, TMEM98, NECTIN3, EDNRA, IL33, FAT4, ACVRL1, HGF, JAM2, ALPL, CD163, SLAMF8, STXBP5, FHL2, ITGA5, DLG4, CRYBG3, TGFBR3, DNAJB4, PRKD1, C7, TMEM200A, CCL19, PDLIM2, ITGAV, RUNX1, YAP1, SPATS2L, C1RL, DZIP1L, PLD1, LATS2, CDH23, C1QTNF6, CD302, CXCL9, F13A1, FOXO1, SOX5
stromal | negative: ABCB1, ABCC12, ABCC8, ABI3, AC007906.2, ACPP, ADAMDEC1, ADCYAP1, ADGRB1, ADGRG6, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADTRP, AGMAT, AIM2, ANGPT1, ANXA3, ANXA8, APLNR, APOBEC3D, ARG2, ARL4D, AUNIP, AURKB, AVPR1A, BANK1, BCL11B, BCL2A1, BCL6B, BLK, BLM, BMP2, BMP6, BMPR1B, BPIFB1, BRCA1, BRCA2, BRINP1, C4B, CABP4, CABYR, CACNG4, CADM3, CALB2, CALML3, CARD9, CASP10, CAVIN2, CCDC141, CCL14, CCL22, CCL28, CCN3, CCNE1, CCR1, CCR4, CCR5, CCR6, CD19, CD1C, CD1D, CD1E, CD22, CD226, CD247, CD27, CD274, CD28, CD33, CD38, CD3G, CD40LG, CD5, CD7, CD72, CD79A, CD79B, CD86, CD8A, CDCA2, CDH13, CDH6, CDKN3, CENPV, CFC1, CHST8, CLDN5, CLEC12A, CLEC14A, CLEC4C, CLEC5A, CMA1, CNR2, COCH, CR1, CRB2, CST7, CTLA4, CTSG, CTSW, CUBN, CUX2, CX3CR1, CXCL2, CXCR3, CXCR6, CYSLTR1, DDX28, DHODH, DLEC1, DLL4, DLX4, DMD, DNAH10, DNAH5, DNASE1L3, DNMT3B, DOC2A, DOK7, DPY19L2, DRC3, DSC3, DSG3, ECRG4, EDN1, EGF, ENPP3, EPHA4, EPHB1, EREG, ERG, ERP27, ERVMER34-1, ESM1, F2RL3, F7, FAM107A, FAM117B, FANCD2, FAT2, FBXO16, FCER1A, FCGR2B, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGFR4, FGR, FLI1, FLT1, FLT3, FLT4, FOLH1, FOXC1, FOXC2, FOXD2, FPR1, FUT7, GABRD, GABRP, GIMAP8, GLS2, GPER1, GPR174, GPR39, GPR4, GRIK4, GRM4, GRP, GUCY1A2, GZMA, GZMB, GZMK, HAVCR2, HBEGF, HCK, HCN2, HDC, HEY1, HMGA1, HOXA5, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HS3ST3A1, ICAM2, ICOS, IDO1, IL12RB1, IL18, IL18R1, IL1RAP, IL1RL1, IL21R, IL2RB, IL3RA, IL6R, IRF4, IRF5, IRX1, ITGA7, ITGB1BP2, ITK, ITPKA, KCNA3, KCNAB3, KCNF1, KCNH6, KCNK10, KCNK17, KCNMB1, KCNN3, KCNN4, KDR, KIT, KLHL13, KLK10, KLK6, KLRD1, KLRG1, KLRK1, LAMB3, LAMP3, LCK, LCN2, LDB3, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LPAR5, LRG1, LTK, MALL, MAP2, MC1R, MECOM, MED12L, MELTF, MET, MLXIPL, MMRN2, MNDA, MRM3, MS4A1, MS4A2, MTNR1B, MUC16, NCCRP1, NCF1, NEIL3, NEK2, NEK8, NIBAN3, NLGN3, NLRP3, NMB, NOD2, NOS1AP, NOS3, NOSTRIN, NPNT, NPY1R, NR4A3, NR5A2, NRROS, NSUN5, NTRK2, NXT2, OLR1, OPRD1, OSBP2, OSGIN1, OXTR, P2RX1, P2RX5, P2RY13, P2RY14, P2RY6, PALMD, PAX5, PCDH10, PCDH17, PCDH8, PDE2A, PDE3A, PDZK1, PGR, PHEX, PHF21B, PILRA, PIMREG, PKP2, PLA2G7, PLK4, POTEI, POTEJ, POU2AF1, POU2F2, PPFIA3, PPP1R1C, PRF1, PRKCQ, PRODH, PROM1, PROX1, PSRC1, PTAFR, PTCH1, PTGDR, PTGS1, PTPRB, QPCTL, RAD54L, RASGRP1, RBBP4, RFX2, RGS5, RIMS3, RIPK3, RNF39, ROBO4, RPA3, RTN1, S1PR1, SAC3D1, SCGB1D2, SCIMP, SCN9A, SDHAF3, SDK2, SELP, SERPINB5, SESN2, SH2D1A, SH2D2A, SH3RF2, SHANK3, SHD, SHLD1, SIGLEC6, SIGLEC9, SIM2, SKA3, SLAMF1, SLAMF6, SLC11A1, SLC16A9, SLC18A2, SLC6A1, SLC7A5, SLCO2A1, SMPD3, SOSTDC1, SOX10, SOX17, SPAG1, SPIB, STAC3, STAP1, STRIP2, STX11, SUV39H1, SYNCRIP, TAL1, TBXA2R, TCL1A, TDO2, TEK, TEKT3, TEX30, TFAP2B, TFR2, TH, TIE1, TIGIT, TLE6, TLR7, TLR9, TMEM255B, TMPRSS13, TNFRSF10C, TNFRSF13B, TNFRSF13C, TNFRSF17, TNFSF15, TNFSF8, TP63, TPSG1, TREM2, TREML2, TRH, TRPC6, TSPAN7, TUBB2B, UBASH3A, UCN, VSNL1, VTCN1, WFDC1, WNT5B, ZBP1, ZFAT, ZNF165, ZNF232, ZNF683
tumor | positive: ECM1, CDH2, ITPRID2, NRAS, FIBCD1, CYP24A1, GSR, TMEM97, CCDC170, FAR1, BMI1, PLA2G6, YJEFN3, MLF1, FMO5, ADGRB2, ITGA2B, CLGN, MTNR1B, DLK1, CENPV, MLXIPL, ABCC12, ERVMER34-1, PHF21B, GRIK4, PPP1R1C, GRM4, DLEC1, PBX1, IKZF2, TBX3, HSPD1, MYO1C, DNMT3A, IARS2, VAPB, CBX5, CDK8, MSH6, API5, REPS1, IPO9, GLUD1, BRI3BP, C1QBP, CPT1A, PPP3CA, PHF8, POLE, SLC25A11, LDLRAD4, PVR, SOX2, UBE2T, NAT10, FH, GSDMB, ZNF687, CACNA1D, STMN1, AURKA, SLC25A40, SPAG5, CENPF, CEP85, GSTZ1, CACNA1B, HS6ST2, ANKS6, WDHD1, PBK, CIP2A, AURKB, CDCA2, NEK2, UCN, DNAH5, MSTN, GPR39, HCN2, FGFR4, F7, SPAG17, PRODH, EGF, BLM, HMGN5, GYG2, KCNH6, SIM2, FBXO16, DNAH10, PIMREG, POTEJ, ABCC8, PCDH8, CORO1B, KIF5B, CDK4, ACKR3, IDUA, ADIPOR1, SMC4, EEF1AKNMT, URB1, DNAJA1, HADHB, AKT2, GFPT1, DDX1, HBS1L, GMPS, KDM6B, POLDIP2, TNFRSF6B, DHX9, CDK16, SHLD2, GPN1, MPDU1, FANCI, PNP, ABCB6, XPO1, NSD2, HSPA4, PPP1R15B, EIPR1, FDFT1, HMGB2, RPA1, DOT1L, HELLS, PELP1, MCM6, POLH, RITA1, UNC5A, TRPV1, RBBP5, TFPI2, HSD11B2, RFC5, SHMT2, CYCS, HCN3, GNAO1, CDKN3, GXYLT2, PLK4, RAD54L, BRCA1, CDC25A, SUV39H1, STAC3, EFR3B, PRDM6, TEX30, CBS, CCNE1, GLS2, AUNIP, ITPKA, TFR2, TEKT3, NLGN3, TH, CHST8, CTTN, COMT, CD46, KDM5B, PPM1H, TOM1L2, HADHA, CCDC14, CKAP5, CTDSPL, BCCIP, PRRC2A, ELMOD2, ACACA, CBX3, SLC39A9, TSC2, GIGYF1, PAPSS1, CLCN3, UFC1, FADD, MSH2, APTX, CISD2, SLC25A1, GHITM, PSMD4, SFXN1, DKC1, SF3B4, HPRT1, SF3B3, CRIPT, LONP1, CARM1, CD320, SNAP47, NONO, MTCH2, SLC20A2, COPS5, SLC4A1AP, THUMPD3, APEX1, DGAT1, TRAP1, PFKP, DHX34, ADIPOR2, FXR2, RAVER1, RCN2, TRIM28, CLK2, DHX57, NEK4, E2F3, CNIH4, RAD18, TRUB1, EHMT2, USP5, NOA1, CNNM3, DVL2, ERCC2, PSEN2, FANCC, DNAJC9, EPRS, FN3KRP, RBBP7, RRS1, CCT2, LSM12, NOP2, TFAM, ZNF281, MIPOL1, MTHFD1, TBXAS1, WDR12, RRM1, CCDC137, NIFK, HGH1, ILF3, CKAP2, MYBL2, PRPS2, ZGRF1, GADD45G, POMT1, HMGA1, MZF1, CABP4, PRRT2, NMB, PSRC1, ABTB2, ERG, CDC6, SESN2, OSCP1, MED12L, AHRR, STEAP1, KITLG, DNMT3B, RPA3, NEIL3, SKA3, ITGB1BP2, STRIP2, TNFRSF10C, DRC3, CRYBG1, NAAA, MPHOSPH9, KMO, AGMAT, FUT8, RFC4, SEMA4C, CHD3, DMXL2, DVL3, DAG1, ZFHX3, SSRP1, MAP4K3, STRN4, KHSRP, NCSTN, PSMB5, GUSB, UBAP2L, MCCC1, PPID, CYHR1, HECTD4, PREPL, IDH1, TMEM101, EIF2AK2, FDPS, GCN1, CTNNAL1, MRFAP1L1, USP10, HSF1, NCLN, SCRN1, MCM7, KRTCAP2, GPBP1, DDX55, HDAC2, ERGIC2, MEIS1, YY1AP1, AIMP2, GART, RO60, TMEM209, NTPCR, IDE, SEPTIN9, ACLY, HDAC4, SLC39A11, SIGMAR1, SLC39A8, TBRG4, PHB2, TMEM120B, TMEM192, MAP2K4, MID2, POLD1, SHMT1, SLC37A4, REEP4, UBAP2, FAM98A, EGLN3, FBXO25, BLOC1S2, BARD1, BCL2L12, IKZF4, RAPGEF3, MRM3, NAIF1, ZNF232, SAC3D1, STS, ZNF408, PRMT5, PRKD1, SYNCRIP, NBPF3, MNS1, OXTR, OSBP2, COLQ, MC1R, ADGRG6, TLE6, ADAM8, LIPE, NEK8, SMARCC1, MYBBP1A, ZNF827, METTL18, QPCTL, AGAP1, SLC29A1, RXRA, PWWP3A, EIF4A1, YES1, PDIA3, ARHGAP35, LGALS8, FHOD1, CRK, INPPL1, KEAP1, IRF2BPL, MTA1, CREG1, TMEM123, POLR2A, SPTBN1, EIF4A3, TMUB2, EMC4, EIF2S1, PKD1, PGD, NUCB2, PTEN, ALKBH5, DHX30, DNMT1, ALDH3A2, BAG3, SLC11A2, SEC13, SHARPIN, RBCK1, FIBP, LTB4R, OCRL, SNX27, RCOR1, DAZAP1, PIGF, RBM10, RAB11FIP1, PRKDC, DAP3, GTPBP4, MEPCE, UBTF, FBL, PRPF31, CCAR2, ELOC, PHF23, MTERF2, APEX2, MRTFB, GSK3A, URGCP, SMC3, TOR1A, RUVBL1, SAT2, CFAP20, MCM3, ECSIT, FECH, RRP15, ERLIN2, NUDT1, RNF121, RWDD4, TUBG1, WDR55, HAX1, CSE1L, LANCL2, BTRC, UMPS, TSPYL5, R3HDM1, ALDH1B1, MAPK12, GMFB, CDAN1, PHLPP1, PTPN11, AMOT, TRAF3IP2, GLOD4, PGAP1, AMACR, GPRASP2, SULT1A1, SSX2IP, ATAD3A, LTBP1, DDX28, DHODH, C5AR1, MECOM, PLIN1, MALL, NPY1R, CABYR, RBM38, NT5E, TRIB3, TP53, POLR2B, SBNO1, PIH1D1, CTCF, PHF12, TOR3A, HNRNPM, ATPSCKMT, PARP1, PNPT1, DDX31, STK16, MANEA, OXSM, FANCD2, FAM117B, BRCA2, KCNAB3, RHOH, ATG16L1
tumor | negative: ABCA10, ABCA8, ABCC9, ABI3, ABI3BP, ACSL4, ACVRL1, ADA, ADAM12, ADAM33, ADAMDEC1, ADAMTS14, ADAMTS4, ADAMTS5, ADCYAP1, ADGRB1, ADGRE2, ADGRL4, ADORA2B, ADORA3, ADRA1B, ADRA2A, ADTRP, AIF1, AIM2, AKR1C3, AKT3, ALPL, ANGPT1, ANGPTL2, ANKRD6, ANXA3, ANXA8, AOAH, APLNR, APOBEC3C, APOBEC3G, APOL3, ASPN, ATL1, AVPR1A, BANK1, BCL11B, BCL2A1, BCL6B, BDKRB2, BLK, BMP2, BMP6, BMPR1B, BNC2, BPIFB1, BRINP1, C1QTNF1, C3AR1, C7, CACNG4, CADM3, CALB2, CALCRL, CALML3, CARD9, CASP1, CASP10, CAVIN2, CCDC136, CCDC141, CCL14, CCL19, CCL2, CCL22, CCN3, CCN4, CCND2, CCR1, CCR4, CCR5, CCR6, CD19, CD1C, CD1D, CD1E, CD209, CD22, CD226, CD247, CD27, CD274, CD28, CD302, CD33, CD38, CD3D, CD3E, CD3G, CD40LG, CD5, CD7, CD72, CD79A, CD79B, CD86, CD8A, CDH13, CDH23, CDH5, CDH6, CDKL1, CEP112, CES1, CLDN5, CLEC10A, CLEC12A, CLEC14A, CLEC4C, CLEC5A, CLEC7A, CMA1, CMKLR1, CNN1, CNR2, CNRIP1, CNTNAP1, COCH, COL10A1, COL4A3, COL4A4, COMP, CPVL, CPXM2, CR1, CRB2, CREB5, CRYBG3, CSF2RA, CSF3R, CSPG4, CST7, CTLA4, CTSG, CTSW, CUBN, CUX2, CX3CL1, CX3CR1, CXCL2, CXCL9, CXCR3, CXCR6, CYSLTR1, DCLK1, DERL3, DIO2, DLL4, DMD, DNASE1L3, DOC2A, DPT, DPY19L2, DSC3, DSG3, DTX1, EBF2, ECRG4, EDN1, EDNRA, EGR2, ENOX1, ENPP3, EPHA4, EPHB1, EREG, ESM1, ETV1, ETV5, F13A1, F2RL3, FAM107A, FAP, FAT2, FAT4, FBLN2, FCER1A, FCGR2A, FCGR2B, FCGR3A, FCMR, FCRL1, FCRL2, FCRL3, FCRL5, FCRLA, FGF1, FGF2, FGF7, FGR, FLI1, FLT1, FLT3LG, FLT4, FMOD, FOLH1, FOLR2, FOXC2, FPR1, FREM1, FST, FUT7, FYN, GAB2, GABRD, GABRP, GBP5, GIMAP8, GLI2, GPR174, GPR34, GPR4, GRP, GUCY1A2, GZMA, GZMB, GZMK, HAS2, HAVCR2, HBEGF, HCK, HDC, HEYL, HGF, HLX, HMCN1, HOXA5, HOXB4, HOXD10, HOXD9, HPGD, HPGDS, HPSE, HPSE2, HS3ST3A1, HSPB8, ICAM2, IDO1, IFI44L, IFITM1, IGF1, IL12RB1, IL18, IL18R1, IL1RL1, IL21R, IL2RB, IL33, IL34, IL3RA, IL6R, INHBA, IRF4, IRF5, IRX1, ITIH5, ITK, JAM2, KCNA3, KCNF1, KCNJ8, KCNK10, KCNK17, KCNMB1, KCNN3, KCNN4, KDR, KIAA0408, KIT, KLHL13, KLK10, KLK6, KLRD1, KLRG1, KLRK1, LACC1, LAIR1, LAMP3, LCK, LCN2, LCNL1, LDB3, LIF, LILRA4, LILRB2, LILRB4, LILRB5, LOX, LPAR1, LPL, LRRC15, LTK, LURAP1, LY96, LZTS1, MAP1A, MAP3K7CL, MAPK8IP1, MELTF, MERTK, MFAP5, MITF, MMP9, MNDA, MPP1, MRC1, MS4A1, MS4A2, MSR1, MUC16, NAV2, NCCRP1, NCF1, NCF2, NEXN, NFATC1, NGFR, NIBAN3, NLRP3, NOD2, NOS3, NPNT, NPR1, NR4A3, NR5A2, NRN1, NRROS, NTRK2, OLFML3, OLR1, OPRD1, P2RX1, P2RX5, P2RX7, P2RY13, P2RY14, P2RY6, PALMD, PAMR1, PAX5, PCDH10, PCDH17, PCDH18, PDE2A, PDE3A, PDGFRA, PGF, PGR, PHEX, PI16, PIEZO2, PLA2G7, PLEKHH2, POU2AF1, POU2F2, PPFIA3, PRF1, PRICKLE1, PRKCQ, PROCR, PROM1, PROS1, PROX1, PTAFR, PTGDR, PTGIR, PTGIS, PTGS1, PTPRB, PTPRM, RASGRP1, RCAN2, RECK, RELN, RGS5, RIMS3, RNF180, RNF39, ROBO4, ROR2, RTN1, RUNX1T1, RUNX2, S1PR1, SAMD5, SATB1, SCGB1D2, SCIMP, SCN9A, SDHAF3, SDK2, SELP, SEMA7A, SERPINB5, SFRP4, SH2D1A, SHD, SIGLEC1, SIGLEC6, SIGLEC9, SLAMF1, SLAMF6, SLAMF8, SLC11A1, SLC18A2, SLC1A3, SLC2A3, SLC35G2, SLC6A1, SLC7A7, SLCO2A1, SLCO2B1, SLIT2, SMPD3, SOCS2, SOD3, SOSTDC1, SOX10, SOX17, SOX5, SPIB, SPOCK1, SPON1, SRPX, STAP1, STAT4, STAT5A, STMN3, STX11, SUGCT, SVEP1, TAL1, TBX2, TBXA2R, TCF7, TCL1A, TDO2, TEK, TFAP2B, TGFB2, TGFBR3, TGM2, THBD, THEMIS, TIE1, TIGIT, TLR1, TLR2, TLR7, TLR9, TMEM119, TMEM200A, TMEM255B, TMEM98, TMPRSS13, TNFRSF13B, TNFRSF17, TNFSF15, TNFSF8, TP63, TPSG1, TRBC1, TREM2, TREML2, TRH, TRPC1, TRPC6, TSPAN18, TSPAN7, TUBB2B, TWIST1, TWIST2, UBASH3A, VGLL3, VSIG4, VSNL1, VTCN1, WFDC1, WNT2, WNT4, WNT5B, ZAP70, ZBP1, ZFHX4, ZNF608, ZNF683
In such cases, the marker definition can be relaxed by adjusting the thresholds used in markers_from_reference above.
[11]:
ctypes = list(markers.keys())
overlap_df = pd.DataFrame(0, index=ctypes, columns=ctypes, dtype=int)
for c in ctypes:
neg_c = set(markers[c].get("negative", []))
for d in ctypes:
pos_d = set(markers[d].get("positive", []))
overlap_df.loc[c, d] = len(neg_c & pos_d)
overlap_df
[11]:
| B | DCIS1 | DCIS2 | T | dendritic | endo | macro | mast | myoepi | perivas | stromal | tumor | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | 0 | 58 | 74 | 25 | 43 | 128 | 89 | 23 | 139 | 53 | 101 | 119 |
| DCIS1 | 45 | 0 | 16 | 47 | 59 | 98 | 79 | 26 | 44 | 42 | 66 | 26 |
| DCIS2 | 43 | 11 | 0 | 50 | 58 | 102 | 82 | 25 | 63 | 44 | 73 | 5 |
| T | 34 | 137 | 156 | 0 | 70 | 149 | 132 | 28 | 157 | 61 | 115 | 269 |
| dendritic | 27 | 55 | 76 | 28 | 0 | 115 | 59 | 19 | 123 | 47 | 84 | 127 |
| endo | 41 | 43 | 43 | 38 | 43 | 0 | 73 | 22 | 60 | 4 | 31 | 76 |
| macro | 25 | 48 | 61 | 17 | 32 | 104 | 0 | 16 | 99 | 50 | 78 | 99 |
| mast | 35 | 148 | 172 | 38 | 65 | 127 | 99 | 0 | 124 | 42 | 96 | 298 |
| myoepi | 41 | 8 | 4 | 33 | 47 | 59 | 47 | 18 | 0 | 27 | 37 | 22 |
| perivas | 45 | 50 | 55 | 30 | 48 | 51 | 91 | 25 | 75 | 0 | 41 | 89 |
| stromal | 44 | 43 | 42 | 47 | 48 | 65 | 57 | 24 | 63 | 20 | 0 | 83 |
| tumor | 47 | 21 | 9 | 58 | 62 | 109 | 100 | 27 | 91 | 45 | 82 | 0 |
Compute marker purity#
We next quantify how pure each spatial cell’s expression profile is with respect to its assigned cell type using positive and negative marker genes.
We assume that differences between scRNA-seq and spatial transcriptomics profiles arise primarily from local contamination by neighboring cells (e.g. diffusion or segmentation spillover). This motivates evaluating negative markers in a neighborhood-aware manner.
c, we compute per-cell precision, recall, and F1 based on markers[c]["positive"]. These scores measure how consistently a cell expresses markers expected for its assigned type. Marker presence is defined either by a rank-based quantile rule (use_quantiles=True, default) or by expression > 0.require_neighbor_expression = True). These relevant negative markers represent genes that should be absent from the focal cell but present in nearby cells.
Precision, recall, and F1 then quantify how strongly the focal cell avoids expressing neighborhood-associated markers.If no neighborhood graph is present (neighbors_key not in sdata.tables[tables_key].obsp), it is automatically computed from cell centroids using a Delaunay graph.
F1_purity score, optionally with a weighting factor for negative_F1 (weight_cont, default=2.0). High purity indicates strong expression of cell-type–specific markers and minimal expression of markers attributable to neighboring cells.All per-cell metrics are returned and, if inplace=True, stored in sdata.tables[tables_key].obs.
[12]:
purity = st.sp.marker_purity(
cell_type_key="transferred_cell_type",
markers=markers,
weight_cont=0.7,
require_neighbor_expression=True,
neighbors_key="spatial_connectivities",
use_quantiles=False,
inplace=True,
)
INFO Creating graph using `generic` coordinates and `None` transform and `1` libraries.
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/src/segtraq/SegTraQ.py:1195: RuntimeWarning: neighbors_key='spatial_connectivities' not found in adata.obsp. A neighborhood graph based on Delaunay will be computed.
return sp.marker_purity(
negative_F1#
[13]:
def boxplot_per_celltype(
sdata,
feature,
celltype_col="transferred_cell_type",
q=1.0,
figsize=(10, 5),
palette=None,
):
obs = sdata.tables["table"].obs
df = obs[[celltype_col, feature]].dropna().copy()
if q < 1:
df = df[df[feature] <= df[feature].quantile(q)]
# order by median (low -> high). Use .sort_values(ascending=False) for high -> low
order = df.groupby(celltype_col)[feature].median().sort_values().index.tolist()
# ensure palette covers all categories (optional safety)
if palette is not None:
palette = {ct: palette.get(ct, "#808080") for ct in order}
fig, ax = plt.subplots(figsize=figsize)
sns.boxplot(
data=df,
x=celltype_col,
y=feature,
order=order,
hue=celltype_col,
palette=palette,
showcaps=True,
showfliers=False,
legend=False,
ax=ax,
)
sns.stripplot(
data=df,
x=celltype_col,
y=feature,
order=order,
hue=celltype_col,
palette=palette,
jitter=0.25,
dodge=False,
alpha=0.6,
edgecolor="black",
linewidth=0.4,
legend=False,
ax=ax,
)
ax.tick_params(axis="x", rotation=45)
ax.set_xlabel("Cell type")
ax.set_ylabel(feature)
fig.tight_layout()
plt.show()
The negative_F1 score indicates that cells of type DCIS1 are the most contaminated.
[14]:
boxplot_per_celltype(st.sdata, feature="negative_F1", palette=col_celltype)
/tmp/ipykernel_27337/3436113034.py:16: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
order = df.groupby(celltype_col)[feature].median().sort_values().index.tolist()
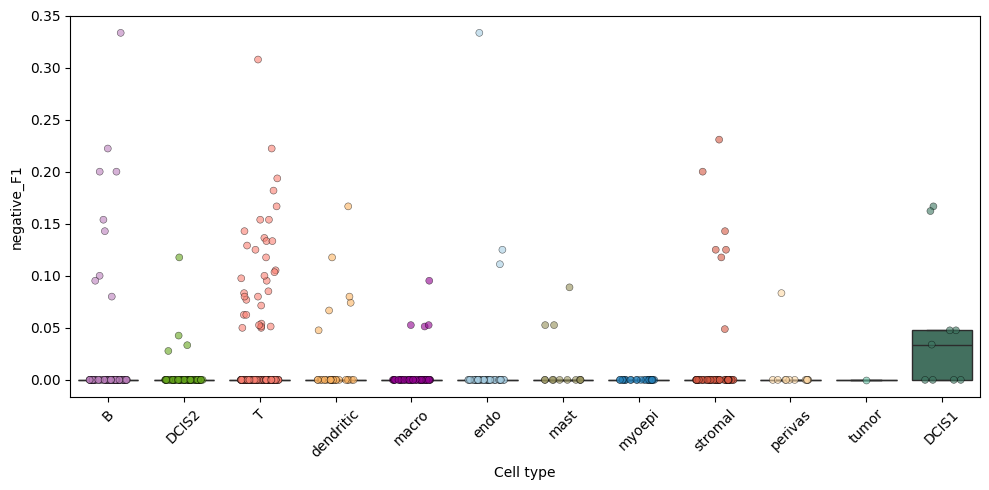
This can also be visualized spatially. The negative_F1 score is only computed for cells which have a neigbhor of a different type. This clearly shows the DCIS1 cells with high negative_F1 score, i.e. contamination.
[15]:
axes = plt.subplots(1, 2, figsize=(10, 5), constrained_layout=True)[1].flatten()
st.sdata.pl.render_shapes(
"cell_boundaries",
color="transferred_cell_type_plot",
palette=cols,
groups=labels,
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[0], title="Cell boundaries colored by cell type", coordinate_systems="global")
st.sdata.pl.render_shapes(
"cell_boundaries",
color="negative_F1",
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[1], title="Cell boundaries colored by negative_F1", coordinate_systems="global")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata_plot/pl/utils.py:872: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = color_source_vector.map(color_mapping)
WARNING Found 177 NaN values in color data. These observations will be colored with the 'na_color'.
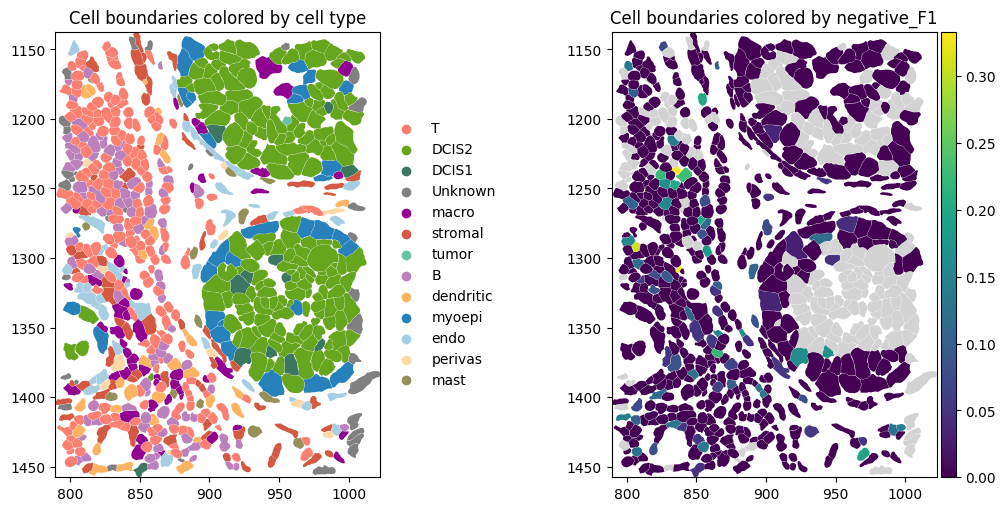
positive_F1#
Cells annotated as mast and DCIS2 show the highest positive_F1 scores, suggesting that their cell identity is captured most consistently by positive markers.
[16]:
boxplot_per_celltype(st.sdata, feature="positive_F1", palette=col_celltype)
/tmp/ipykernel_27337/3436113034.py:16: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
order = df.groupby(celltype_col)[feature].median().sort_values().index.tolist()
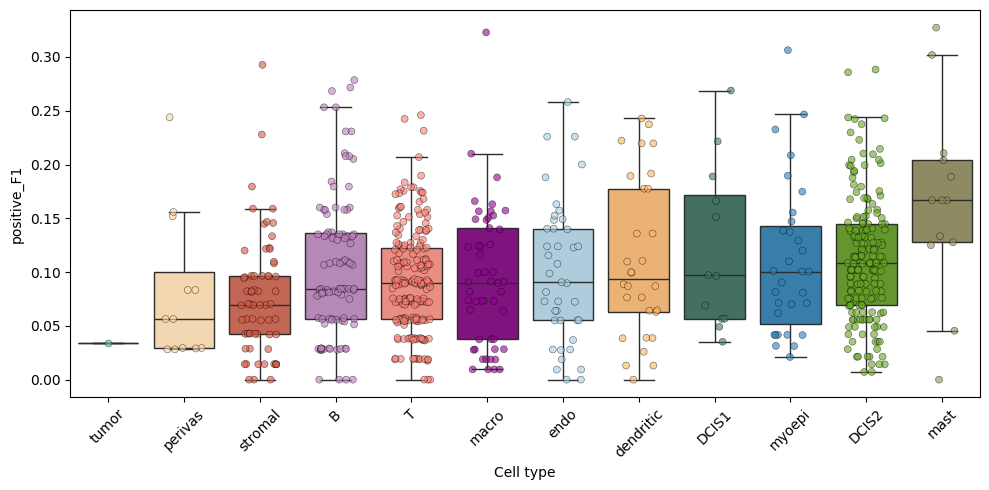
The positive_F1score is computed globally, i.e. irrespective of the cell neighborhood, so there are fewer missing values than for negative_F1. The spatial plot below is in line with the boxplot above, showing that cells of type DCIS2 and mast show the highest positive_F1.
[17]:
axes = plt.subplots(1, 2, figsize=(10, 5), constrained_layout=True)[1].flatten()
st.sdata.pl.render_shapes(
"cell_boundaries",
color="transferred_cell_type_plot",
palette=cols,
groups=labels,
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[0], title="Cell boundaries colored by cell type", coordinate_systems="global")
st.sdata.pl.render_shapes(
"cell_boundaries",
color="positive_F1",
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[1], title="Cell boundaries colored by positive_F1", coordinate_systems="global")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata_plot/pl/utils.py:872: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = color_source_vector.map(color_mapping)
WARNING Found 34 NaN values in color data. These observations will be colored with the 'na_color'.
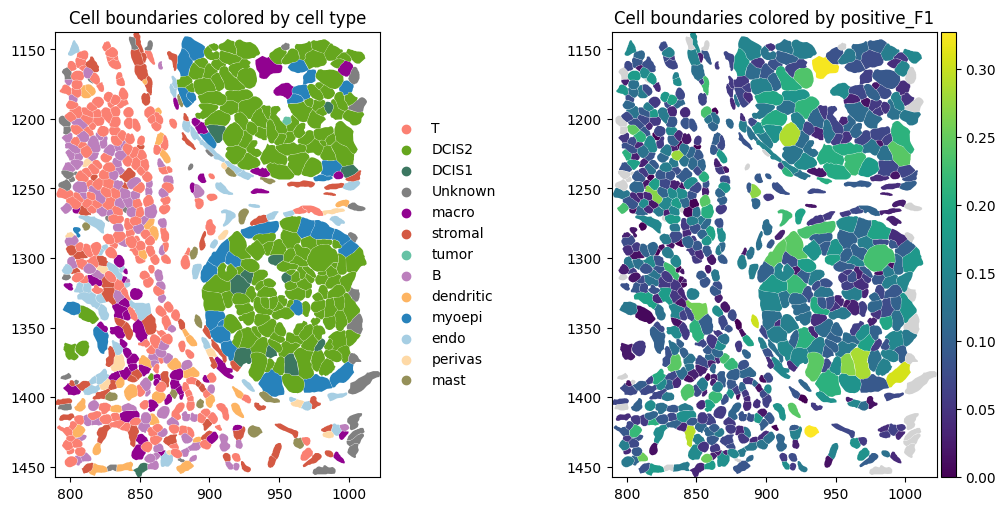
F1_purity#
Cells of type DCIS1 (which have high transcript counts) show the highest negative_F1 and the fourth-highest positive_F1. This is expected: cells with higher transcript counts tend to capture more positive markers, but also have a higher risk of capturing negative markers. As a result, the combined metric F1_purity, which balances positive_F1 and negative_F1, provides a more representative measure of purity. Accordingly, DCIS1 ranks among the cell types with the lowest
purity, while DCIS2 and mast cells show the highest purity.
[18]:
boxplot_per_celltype(st.sdata, feature="F1_purity", palette=col_celltype)
/tmp/ipykernel_27337/3436113034.py:16: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
order = df.groupby(celltype_col)[feature].median().sort_values().index.tolist()
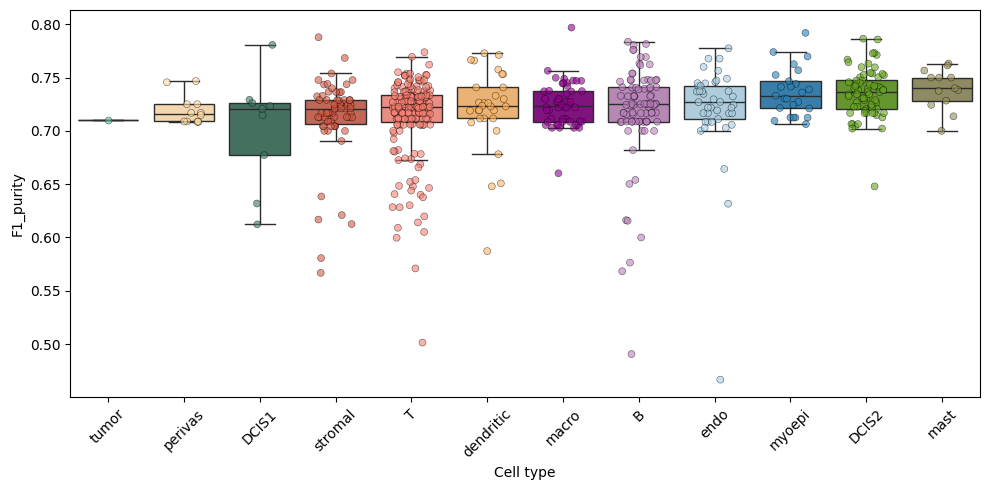
Neighborhood contamination#
Marker purity summarizes how well a cell matches its own markers and avoids neighborhood-relevant negatives. In many cases, we also want to quantify (i) how many contaminating transcripts are present per cell and (ii) which neighboring cell types contribute to this signal. We therefore compute neighborhood contamination.
We treat a gene as contamination-relevant for a target cell type c_tgt if it is (i) a negative marker of ``c_tgt``, (ii) simultaneously a positive marker of a neighboring source type ``c_src``, and (iii) actually expressed (>0) in at least one of the neighboring cells of the source type (require_neighbor_expression , default=True).
This yields two levels of output:
Per-cell metrics. For each cell, we compute
neg_marker_contam_counts: the total number of transcripts for contamination-relevant genes, andneg_marker_contam_fraction: a gene-wise average ofx_i(g) / (x_i(g) + mean_neighbor(g)), which measures how much of the shared marker signal is assigned to the focal cell rather than to its neighborhood.
Directed cell-type contamination (stored in ``.uns``): By aggregating across cells, we construct
uns_key: a directed source → target matrix where each entry is the average strength of contamination from one cell type into another, anduns_key_binary: a complementary directed matrix that reports what percentage of cells of a target type is contaminated by each source type.
If no neighborhood graph is present (neighbors_key not in sdata.tables[tables_key].obsp), it is automatically computed from cell centroids using a Delaunay graph.
[19]:
per_cell_df, cont_mat, cont_bin = st.sp.neighbor_contamination(
cell_type_key="transferred_cell_type",
markers=markers,
require_neighbor_expression=True,
neighbors_key="spatial_connectivities",
uns_key="negative_marker_contamination",
uns_key_binary="negative_marker_contamination_binary",
inplace=True,
)
neg_marker_contam_counts#
The spatial plot below shows, for each target cell, the number of contaminating transcripts. If desired, this quantity can be normalized by the total number of transcripts per cell. As expected, the plot correlates with the negative_F1score from above.
[20]:
axes = plt.subplots(1, 2, figsize=(10, 5), constrained_layout=True)[1].flatten()
st.sdata.pl.render_shapes(
"cell_boundaries",
color="transferred_cell_type_plot",
palette=cols,
groups=labels,
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[0], title="Cell boundaries colored by cell type", coordinate_systems="global")
st.sdata.pl.render_shapes(
"cell_boundaries",
color="neg_marker_contam_counts",
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[1], title="Cell boundaries colored by #contamination transcripts", coordinate_systems="global")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata_plot/pl/utils.py:872: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = color_source_vector.map(color_mapping)
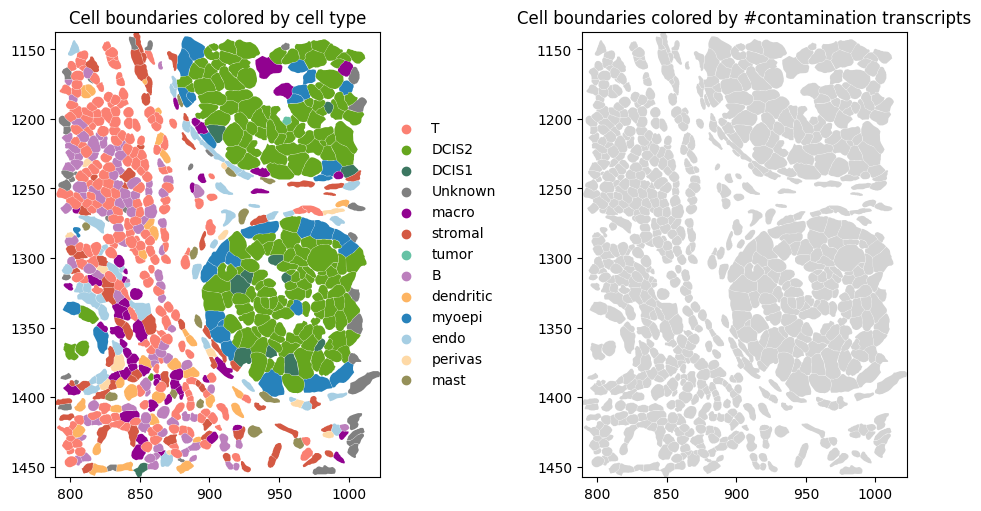
neg_marker_contam_fraction#
In addition, we can plot neg_marker_contam_fraction, the fraction of the signal that is “living” in the wrong cell instead of in the neighbors.
Relevant only if require_neighbor_expression = False: neg_marker_contam_fraction=1 means that 100% of the signal lives in the wrong cell.
[21]:
axes = plt.subplots(1, 2, figsize=(10, 5), constrained_layout=True)[1].flatten()
st.sdata.pl.render_shapes(
"cell_boundaries",
color="transferred_cell_type_plot",
palette=cols,
groups=labels,
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[0], title="Cell boundaries colored by cell type", coordinate_systems="global")
st.sdata.pl.render_shapes(
"cell_boundaries",
color="neg_marker_contam_fraction",
outline_color="white",
outline_width=0.5,
).pl.show(ax=axes[1], title="Cell boundaries colored by contamination fraction", coordinate_systems="global")
/g/huber/users/meyerben/notebooks/spatial_transcriptomics/SegTraQ/.venv/lib/python3.13/site-packages/spatialdata_plot/pl/utils.py:872: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = color_source_vector.map(color_mapping)
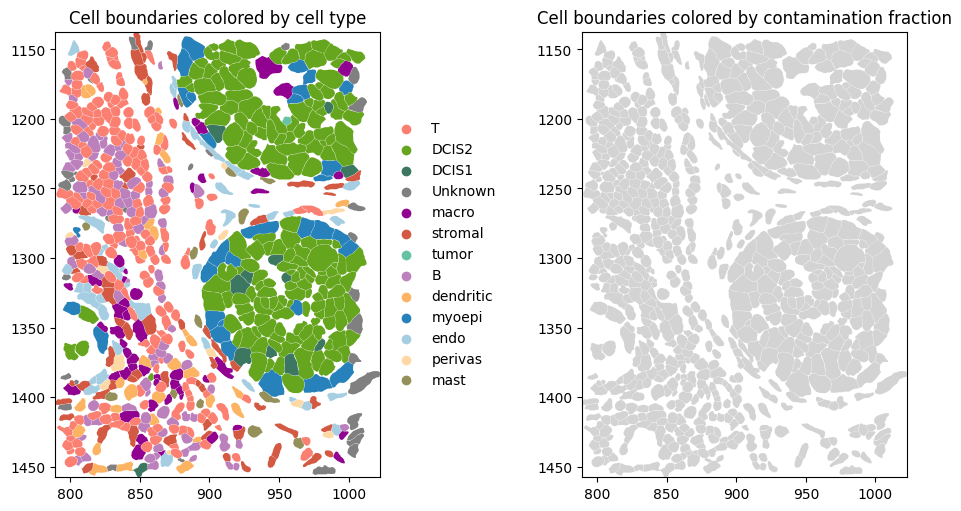
.uns["negative_marker_contamination_binary"]#
The heatmap below shows, for each source–target cell-type pair, the fraction of all target cells contaminated by transcripts from a neighboring source cell type.
Percentages within a column may sum to more than 100% because a single target cell can be contaminated by multiple neighboring source cell types and is therefore counted for each of them.
DCIS1 cells are mostly contaminated by DCIS2 cells, which makes sense since they are frequent neighbors. T cells are contaminated by stromal and B cells.
[22]:
plt.figure(figsize=(8, 6))
sns.heatmap(
cont_bin,
annot=True,
fmt=".2f",
cmap="Reds",
linewidths=0.5,
linecolor="gray", # or "viridis", "magma", "rocket"
)
plt.title("Directed Cell-Type Contamination (c_src → c_tgt)", fontsize=14)
plt.xlabel("Target Cell Type")
plt.ylabel("Source Cell Type")
plt.tight_layout()
plt.show()
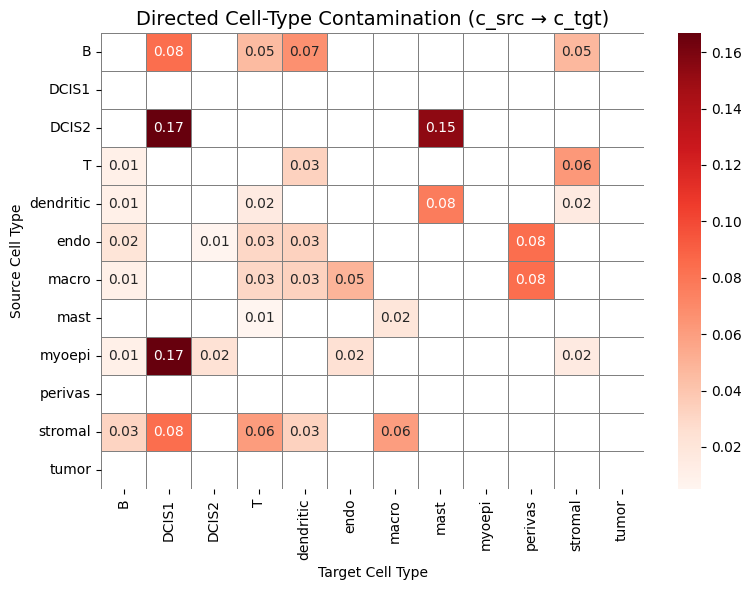
.uns["negative_marker_contamination"]#
Relevant only if require_neighbor_expression = False: Values close to 1 indicate cases where the target cell carries most or all of the detected signal for a given gene, while neighboring source cells show little or no expression of that gene. This typically reflects rare or low-count events, such as single-transcript misassignments.
[23]:
plt.figure(figsize=(8, 6))
sns.heatmap(
cont_mat,
annot=True,
fmt=".2f",
cmap="Reds",
linewidths=0.5,
linecolor="gray", # or "viridis", "magma", "rocket"
)
plt.title("Directed Cell-Type Contamination (c_src → c_tgt)", fontsize=14)
plt.xlabel("Target Cell Type")
plt.ylabel("Source Cell Type")
plt.tight_layout()
plt.show()
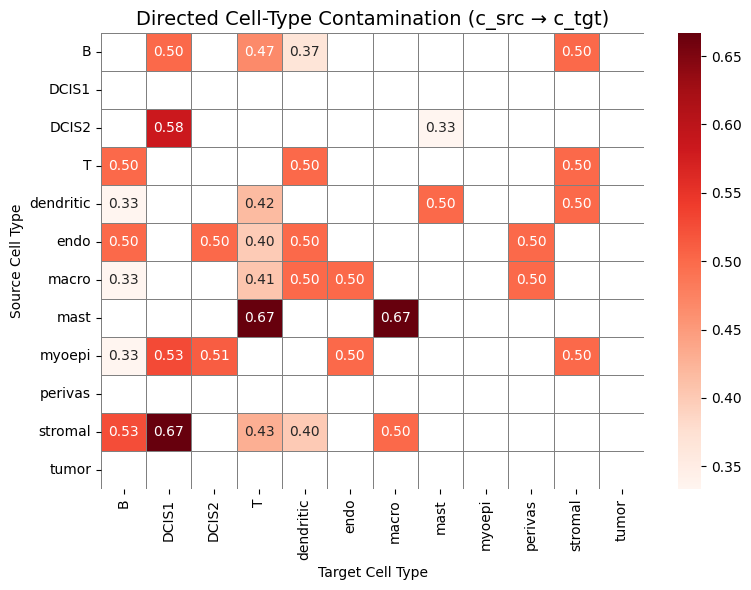
Compute mutually exclusive co-expression rate (MECR)#
The mutually_exclusive_coexpression_rate function assesses mutual exclusivity between marker genes using Fisher’s exact test on binary gene detection (expression > 0). Candidate pairs are constructed as unique, unordered combinations of positive and negative markers across cell types, excluding gene pairs that co-occur as positive markers in any cell type. Fisher’s exact test with alternative="less" is used to test whether genes co-occur less often than expected under independence,
while odds ratios are reported with a pseudocount correction to avoid infinities in sparse data. By conditioning on the marginal detection frequencies of each gene, Fisher’s exact test does not favor methods with low overall transcript counts.
[24]:
mecr = st.sp.mutually_exclusive_coexpression_rate(
markers=markers,
pseudocount=0.5,
inplace=True,
)
[25]:
rows = []
tbl = st.sdata.tables["table"]
mecr_df = tbl.uns["mutually_exclusive_coexpression_rate"]
# Filter significant, finite odds ratios and p-values
df_sig = mecr_df[
mecr_df["odds_ratio"].notna()
& np.isfinite(mecr_df["odds_ratio"])
& mecr_df["pvalue"].notna()
& np.isfinite(mecr_df["pvalue"])
& (mecr_df["pvalue"] < 0.05)
]
# Build rows
rows.extend({"method": "Xenium", "Fisher_OR": np.log2(row["odds_ratio"])} for _, row in df_sig.iterrows())
df = pd.DataFrame(rows)
median_order = df.groupby("method")["Fisher_OR"].median().sort_values().index.tolist()
# Compute medians in plotting order
medians = df.groupby("method")["Fisher_OR"].median().reindex(median_order)
# Build x-axis labels with medians
xtick_labels = [f"{m}\nmedian:({medians[m]:.2f})" for m in median_order]
plt.figure(figsize=(3, 4))
# --- Violin plot ---
ax = sns.violinplot(
data=df,
x="method",
y="Fisher_OR",
order=median_order,
palette="Set2",
linewidth=2,
)
# --- Stripplot overlay ---
sns.stripplot(
data=df,
x="method",
y="Fisher_OR",
order=median_order,
color="black",
size=2.5,
alpha=0.35,
jitter=0.25,
ax=ax,
)
# Reference line: OR = 1
ax.axhline(0, ls="--", lw=1, color="gray", alpha=0.6)
# Apply new x-axis labels
ax.set_xticklabels(xtick_labels)
ax.set_ylabel("log2 Fisher odds ratio")
ax.set_xlabel("")
ax.set_title("Significant associations among candidate mutually exclusive marker pairs")
plt.grid(axis="y", alpha=0.3)
plt.tight_layout()
plt.show()
/tmp/ipykernel_27337/1855350009.py:31: FutureWarning:
Passing `palette` without assigning `hue` is deprecated and will be removed in v0.14.0. Assign the `x` variable to `hue` and set `legend=False` for the same effect.
ax = sns.violinplot(
/tmp/ipykernel_27337/1855350009.py:57: UserWarning: set_ticklabels() should only be used with a fixed number of ticks, i.e. after set_ticks() or using a FixedLocator.
ax.set_xticklabels(xtick_labels)
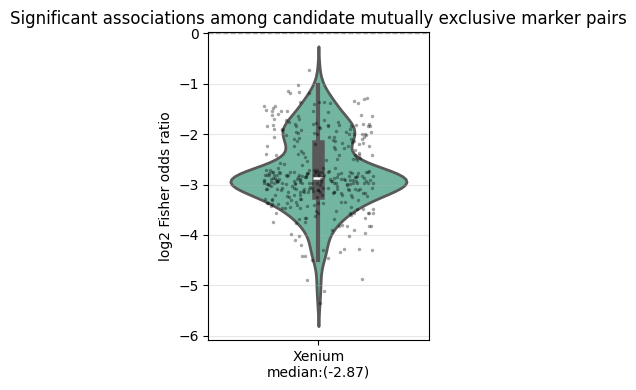
[26]:
print(sd.__version__) # spatialdata
print(spatialdata_plot.__version__)
0.7.2
0.2.13
